writeAlizer: Scoring Model Development
Sterett H. Mercer
Source:vignettes/scoring-model-development.Rmd
scoring-model-development.RmdThis vignette provides details on the scoring models included in writeAlizer.
Recommended Models for Use
ReaderBench-Model-3 and Coh-Metrix-Model-3 are the best models for generated predicted writing quality scores, and aWE-CBM-Model-1 is the best available model for generating automated written expression curriculum-based measurement scores.
Scoring Model Development
The general process used to generate all scoring models is presented below.
Predictive Algorithms and R Packages Used
The caret
and caretEnsemble
packages were used as wrappers for the following predictive
algorithms:
Random forest regression (package
randomForest)Cubist regression (package
Cubist)Support vector machines with a radial kernel (package
kernlab)Bagged multivariate adaptive regression splines (package
earth)Stochastic gradient boosted trees (package
gbm)Partial least squares regression (package
pls)Elasticnet regression (package
elasticnet)
These algorithms are described in detail in the following references (among others):
Hastie, T., Tibshirani, R., & Friedman, J. (2009). The elements of statistical learning: Data mining, inference, and prediction (2nd ed.). Springer. https://doi.org/10.1007/b94608
Kuhn, M., & Johnson, K. (2013). Applied predictive modeling. Springer. https://doi.org/10.1007/978-1-4614-6849-3
Steps
The following flowchart provides an overview of the scoring model development workflow, with more details on some steps provided below.
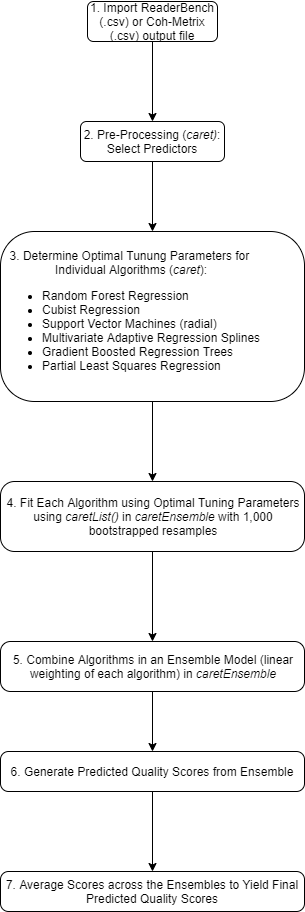
1. Import Data
Depending on the specific scoring model, ReaderBench, Coh-Metrix, and/or GAMET output files were imported into R using functions similar to the import_XXXX.R functions in writeAlizer (see https://shmercer.github.io/writeAlizer/reference/index.html)
2. Pre-Process Data
Automated data pre-processing were done using the preProcess()
function in caret:
Predictors from the output file with near zero variance (defined based on defaults in the
nearZeroVar()function) were removed, and the remaining predictors were standardized.Highly correlated | r > .90 | predictors were identified, with the predictor that had the highest mean correlation with all of the other predictors removed.
The reduced set of predictors was submitted to the next step of the analysis.
3. Determine Optimal Tuning Parameters
The following tuning hyperparameters were optimized based on
resampling (repeated 10 fold) in caret. Each algorithm was
tuned separately. Full descriptions of the tuning parameters are
available in each package’s documentation.
Random forest regression:
mtryCubist regression:
committees,neighborsSupport vector machines:
sigma,CBagged multivariate adaptive regression splines:
nprune,degreeStochastic gradient boosted trees:
n.trees,interaction.depth,shrinkage,n.minobsinnodePartial least squares regression:
ncompElastic net regression:
fraction,lambda
4. Final/Optimal Model for each Algorithm
A model for each algorithm was fit with the hyperparameters set to
the optimal values found in Step 3, with bootstrapped (1000 samples)
resampling-based cross-validation so that an ensemble model (weighting
each algorithm) could be built based on the resamples. This step was
done with the caretList()
function of the caretEnsemble package. This process is
illustrated in more detail in the caretEnsemble vignette:
https://zachmayer.github.io/caretEnsemble/articles/caretEnsemble-intro.html
5. Estimate an Ensemble Model to Combine the Algorithms
The caretEnsemble()
function was used to determine the optimal linear weighting of the
algorithms that minimized RMSE (i.e., discrepancy between actual writing
quality scores and predicted quality scores) in the resamples from Step
4. Algorithms with near zero or negative weights were removed from the
ensemble models.
The varImp()
function of caretEnsemble was used to generate estimates of
relative predictor importance for the overall ensemble model and for
each individual algorithm.
6. Generate Predicted Quality Scores from each Ensemble
The predict()
function of caretEnsemble was used to generate/store
predicted quality scores for the ensemble models.
ReaderBench Model 1
General Description
Model 1 has been replaced by the greatly simplified Model 2 that better handles multi-paragraph compositions. Model 2 is recommended over Model 1.
Model 1 is an ensemble (formed by averaging predicted quality scores) of the six sub-models described below.
All of these sub-models used ReaderBench scores on 7 min
narrative writing samples (“I once had a magic pencil and …”) from
students in the fall, winter, and spring of Grades 2-5 (Mercer et al., 2019) to predict holistic
writing quality on the samples (elo ratings calculated from paired
comparisons). More details on the sample are available in (Mercer et al., 2019).
Highly correlated ReaderBench metrics (r > |.90|) were excluded during pre-processing (see section on Scoring Model Development for more details).
This scoring model was evaluated in the following publications: (Keller-Margulis et al., 2021; Matta et al., 2022; Mercer & Cannon, 2022)
ReaderBench Model 1a
This model was trained on fall data in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -3.9077 | -0.1323 | 0.4789 | -0.0963 | -0.0361 | 0.3985 | 0.1297 | 0.3442 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| WdEnt | 14.61 | 40.66 | 2.49 | 1.93 | 13.17 | 3.97 | 62.12 | 19.56 |
| LxcDiv | 3.02 | 4.3 | 2.2 | 1.52 | 2.69 | 2.77 | 0 | 5.55 |
| AvgUnqPrepositionBl | 2.88 | 3.21 | 2.18 | 1.42 | 6.67 | 1.98 | 0 | 5.84 |
| AvgNmdEntBl | 2.71 | 0.47 | 1.13 | 0.4 | 0.79 | 0.89 | 18.1 | 2.92 |
| AvgUnqVerbBl | 2.4 | 2.36 | 2.21 | 1.48 | 6.53 | 2.33 | 0 | 3.5 |
| WdDiffLemmaStem | 2.07 | 1.07 | 0.82 | 0.91 | 0 | 1.35 | 13 | 1.46 |
| RdbltyFlesch | 1.91 | 0.7 | 0.41 | 0.41 | 4.1 | 1.04 | 6.51 | 3.94 |
| AvgChainSpan | 1.84 | 3.5 | 1.77 | 1.16 | 0 | 2.11 | 0 | 2.04 |
| AvgDepsBl_det | 1.64 | 2.01 | 1.63 | 0.79 | 3.96 | 1.6 | 0 | 2.19 |
| AvgBlScore | 1.55 | 0.87 | 2.01 | 1.32 | 0 | 1.76 | 0 | 1.75 |
| AvgPronBl_first_person | 1.52 | 0.7 | 1.56 | 0.75 | 2.38 | 1.39 | 0 | 2.63 |
| AvgDepsBl_nsubj | 1.5 | 1.34 | 2.18 | 1.45 | 0 | 1.9 | 0 | 0.88 |
| AvgUnqAdverbBl | 1.48 | 1.86 | 1.88 | 1.06 | 1.3 | 2.12 | 0 | 0.73 |
| AvgDepsBl_punct | 1.45 | 1.45 | 1.77 | 0.93 | 1.9 | 2.1 | 0 | 0.88 |
| WdDiffWdStem | 1.43 | 2.02 | 1.47 | 0.84 | 1.36 | 1.04 | 0 | 2.34 |
| AvgDepsSen_punct | 1.41 | 0.64 | 1.33 | 0.65 | 4.76 | 1.04 | 0 | 2.63 |
| AvgDepsSen_dep | 1.34 | 0.04 | 0.65 | 0.33 | 5.78 | 0.53 | 0 | 4.09 |
| AvgSenScore | 1.28 | 0.39 | 0.27 | 0.22 | 0 | 0.53 | 0 | 4.82 |
| AvgPronounBl | 1.23 | 0.2 | 1.8 | 1.08 | 0 | 1.38 | 0 | 1.31 |
| TCorefChainDoc | 1.2 | 0.41 | 1.59 | 0.77 | 0 | 0.88 | 0 | 2.04 |
| AvgDepsBl_nmod | 1.12 | 0.52 | 1.9 | 1.09 | 0 | 1.57 | 0 | 0.29 |
| AvgAOASen_Shock | 1.09 | 1.04 | 0.77 | 0.52 | 3.75 | 1.04 | 0 | 1.9 |
| AvgSenBl | 1.03 | 0.42 | 1.51 | 0.68 | 0 | 1.29 | 0 | 0.88 |
| WdLettStdDev | 1.01 | 0.85 | 1.27 | 0.8 | 0 | 0.84 | 0 | 1.46 |
| AvgWdLen | 0.97 | 1.33 | 1.4 | 0.75 | 0 | 1.26 | 0 | 0.44 |
| AvgDepsBl_nummod | 0.95 | 0.75 | 1.16 | 0.42 | 0.04 | 1.23 | 0 | 1.02 |
| AvgCorefChain | 0.94 | 0.56 | 1.01 | 0.34 | 2.87 | 0.32 | 0 | 2.04 |
| TActCorefChainWd | 0.93 | 0.78 | 0.83 | 0.55 | 0.99 | 1.15 | 0 | 1.31 |
| RdbltyDaleChall | 0.92 | 0.93 | 0.95 | 0.38 | 0 | 1.07 | 0.27 | 1.17 |
| AvgDepsBl_advmod | 0.89 | 0.2 | 1.65 | 0.84 | 1 | 1.26 | 0 | 0 |
| AvgAOABl_Bristol | 0.86 | 0.73 | 0.93 | 0.41 | 0 | 0.52 | 0 | 1.75 |
| AvgUnqNoundBl | 0.85 | 0.28 | 1.64 | 0.82 | 0.65 | 0.88 | 0 | 0.29 |
| AvgDepsBl_dobj | 0.85 | 0.19 | 1.49 | 0.68 | 1.5 | 0.93 | 0 | 0.44 |
| AvgAOABl_Shock | 0.83 | 2.04 | 1.3 | 0.63 | 0.12 | 0.96 | 0 | 0 |
| SenStdDevWd | 0.77 | 0.67 | 1.07 | 0.56 | 0.83 | 1.42 | 0 | 0 |
| AvgDepsBl_mark | 0.76 | 0.15 | 1.31 | 0.53 | 1.23 | 0.7 | 0 | 0.58 |
| LexChainMaxSp | 0.75 | 0.31 | 1.57 | 0.75 | 0 | 0.85 | 0 | 0 |
| WdAvgDpthHypernymTree | 0.75 | 0.42 | 0.63 | 0.36 | 0.57 | 0.6 | 0 | 1.61 |
| AvgPronBl_indefinite | 0.72 | 0.12 | 1.44 | 0.64 | 0 | 0.62 | 0 | 0.44 |
| AvgSenAdjCoh_Path | 0.69 | 0.35 | 0.83 | 0.49 | 0.57 | 0.77 | 0 | 0.88 |
| AvgDepsBl_cop | 0.68 | 0.15 | 1.2 | 0.44 | 2.55 | 0.92 | 0 | 0 |
| CharEnt | 0.68 | 0.26 | 1.44 | 0.69 | 0.42 | 0.73 | 0 | 0 |
| AvgConnBl_simp_subords | 0.66 | 0.11 | 1.2 | 0.45 | 0.31 | 1.05 | 0 | 0 |
| LexChainAvgSpan | 0.66 | 0.49 | 1.17 | 0.7 | 0 | 0.94 | 0 | 0 |
| AvgConnBl_reas_purp | 0.66 | 0.11 | 1.26 | 0.5 | 0 | 1.02 | 0 | 0 |
| AvgDepsBl_advcl | 0.64 | 0.05 | 1.35 | 0.56 | 1.13 | 0.7 | 0 | 0 |
| TCorefChainBigSpan | 0.63 | 0.01 | 1.16 | 0.42 | 1.31 | 0.54 | 0 | 0.44 |
| AvgUnqAdjectiveBl | 0.63 | 0.48 | 1.37 | 0.58 | 0 | 0.61 | 0 | 0 |
| AvgDepsBl_amod | 0.63 | 0.05 | 1.28 | 0.51 | 0.32 | 0.85 | 0 | 0 |
| AvgDepsBl_aux | 0.62 | 0.57 | 0.98 | 0.3 | 0 | 1.07 | 0 | 0 |
| WdPathCntHypernymTree | 0.62 | 0.99 | 0.79 | 0.44 | 3.29 | 0.7 | 0 | 0.15 |
| WdSylCnt | 0.62 | 0.37 | 0.8 | 0.56 | 0 | 1.3 | 0 | 0 |
| AvgUnqPronounBl | 0.61 | 0.21 | 1.3 | 0.53 | 0.62 | 0.65 | 0 | 0 |
| FrqRhythmId | 0.59 | 0.73 | 0.9 | 0.44 | 0 | 0.94 | 0 | 0 |
| AvgDepsBl_nsubjpass | 0.57 | 0.05 | 1.01 | 0.32 | 1.18 | 0.88 | 0 | 0 |
| AvgDepsBl_ccomp | 0.55 | 0.26 | 1.13 | 0.39 | 1.76 | 0.54 | 0 | 0 |
| AvgConnSen_addition | 0.54 | 0.25 | 0.59 | 0.25 | 0 | 0.95 | 0 | 0.44 |
| AvgConnBl_order | 0.52 | 0.07 | 0.72 | 0.16 | 2.74 | 0.92 | 0 | 0 |
| AvgBlVoiceCoOcc | 0.52 | 0 | 1.09 | 0.36 | 0 | 0.73 | 0 | 0 |
| AvgInferenceDistChain | 0.51 | 0.32 | 0.74 | 0.45 | 0.3 | 0.79 | 0 | 0.15 |
| AvgAOESen_InvLinRegSlo | 0.5 | 0.56 | 0.65 | 0.35 | 0.86 | 0.28 | 0 | 0.73 |
| AvgRhythmUnitStreesSyll | 0.49 | 0.29 | 0.03 | 0.3 | 0 | 1.02 | 0 | 0.88 |
| AvgConnBl_semi_coords | 0.48 | 0.01 | 0.98 | 0.3 | 0 | 0.68 | 0 | 0 |
| LxcSoph | 0.48 | 0.25 | 0.83 | 0.32 | 0 | 0.8 | 0 | 0 |
| AvgConnBl_logical_cons | 0.47 | 0.05 | 0.58 | 0.1 | 1.58 | 0.63 | 0 | 0.44 |
| AvgDepsBl_neg | 0.47 | 0.04 | 0.81 | 0.21 | 0 | 0.86 | 0 | 0 |
| AvgCommaBl | 0.47 | 0.09 | 0.9 | 0.25 | 0 | 0.75 | 0 | 0 |
| AvgNounNmdEntBl | 0.47 | 0.08 | 0.67 | 0.14 | 0 | 0.43 | 0 | 0.73 |
| AvgNounSen | 0.46 | 0.23 | 0.12 | 0.07 | 0 | 0.62 | 0 | 1.17 |
| AvgAdverbSen | 0.45 | 0.17 | 0.46 | 0.55 | 0 | 0.81 | 0 | 0.29 |
| AvgConnBl_oppositions | 0.44 | 0.07 | 0.9 | 0.26 | 0 | 0.62 | 0 | 0 |
| AvgNmdEntSen | 0.44 | 0.62 | 0.13 | 0.5 | 0 | 0.93 | 0 | 0.44 |
| AvgConnBl_contrasts | 0.44 | 0.25 | 1.02 | 0.32 | 0 | 0.41 | 0 | 0 |
| AvgConnSen_semi_coords | 0.43 | 0.05 | 0.43 | 0.06 | 0.08 | 0.93 | 0 | 0.29 |
| AvgAOASen_Bristol | 0.43 | 0.48 | 0.32 | 0.48 | 1.36 | 0.7 | 0 | 0.29 |
| AvgDepsBl_xcomp | 0.43 | 0.11 | 1.15 | 0.41 | 0 | 0.23 | 0 | 0 |
| AvgConnSen_simp_subords | 0.43 | 0.74 | 0.28 | 0.31 | 1.49 | 0.97 | 0 | 0 |
| WdPolysemyCnt | 0.41 | 0.4 | 0 | 0.71 | 0.46 | 0.17 | 0 | 1.31 |
| AvgPronBl_third_person | 0.41 | 0.39 | 0.88 | 0.24 | 0 | 0.44 | 0 | 0 |
| AvgAOASen_Bird | 0.4 | 0.25 | 0.58 | 0.51 | 0 | 0.7 | 0 | 0 |
| AvgDepsBl_mwe | 0.4 | 0.03 | 0.49 | 0.08 | 3.31 | 0.71 | 0 | 0 |
| AvgPronounSen | 0.38 | 0.27 | 0.06 | 0.18 | 0 | 0.59 | 0 | 0.88 |
| AvgConnBl_addition | 0.37 | 0.23 | 0.58 | 0.1 | 0 | 0.71 | 0 | 0 |
| AvgAOABl_Kuperman | 0.37 | 0.6 | 0.32 | 0.48 | 0 | 0.43 | 0 | 0.44 |
| AvgAOASen_Kuperman | 0.36 | 0.16 | 0.62 | 0.55 | 0 | 0.52 | 0 | 0 |
| AvgAOABl_Cortese | 0.36 | 0.33 | 0.3 | 0.59 | 2.07 | 0.66 | 0 | 0 |
| AvgDepsSen_advcl | 0.35 | 0.22 | 0.5 | 0.51 | 0 | 0.6 | 0 | 0 |
| AvgDepsSen_det | 0.33 | 0.32 | 0.08 | 0.27 | 0 | 0.31 | 0 | 0.88 |
| AvgDepsBl_acl | 0.33 | 0.01 | 0.54 | 0.1 | 0 | 0.67 | 0 | 0 |
| AvgRhythmUnits | 0.33 | 0.54 | 0.6 | 0.43 | 0 | 0.22 | 0 | 0.15 |
| AggPronSen_indefinite | 0.32 | 0.14 | 0.24 | 0.49 | 0.23 | 0.55 | 0 | 0.29 |
| AvgConnBl_temp_cons | 0.31 | 0.16 | 0.87 | 0.24 | 0 | 0.1 | 0 | 0 |
| AvgDepsSen_ccomp | 0.3 | 0.19 | 0.04 | 0.34 | 0 | 0.79 | 0 | 0.29 |
| SenAsson | 0.29 | 0.02 | 0.43 | 0.06 | 1 | 0.57 | 0 | 0 |
| AggPronSen_third_person | 0.29 | 0.46 | 0.3 | 0.15 | 0 | 0.52 | 0 | 0.15 |
| AvgAOABl_Bird | 0.29 | 0.27 | 0.46 | 0.53 | 0 | 0.3 | 0 | 0.15 |
| AvgDepsSen_aux | 0.29 | 0.08 | 0.12 | 0.49 | 0 | 0.9 | 0 | 0 |
| AvgDepsSen_dobj | 0.28 | 0.1 | 0.11 | 0.06 | 0 | 0.95 | 0 | 0 |
| AvgConnSen_oppositions | 0.26 | 0.03 | 0.28 | 0.03 | 0 | 0.69 | 0 | 0 |
| AvgDepsSen_nmod | 0.26 | 0.18 | 0.37 | 0.37 | 0 | 0.47 | 0 | 0 |
| AvgDepsSen_amod | 0.25 | 0.26 | 0.17 | 0.1 | 0 | 0.47 | 0 | 0.29 |
| AvgAOASen_Cortese | 0.24 | 0.2 | 0.59 | 0.44 | 0 | 0.08 | 0 | 0 |
| AvgDepsSen_compound | 0.24 | 0.16 | 0.38 | 0.18 | 0 | 0.43 | 0 | 0 |
| AvgDepsSen_mark | 0.23 | 0.3 | 0.23 | 0.27 | 0 | 0.48 | 0 | 0 |
| AvgDepsSen_mwe | 0.22 | 0.1 | 0.2 | 0.02 | 0 | 0.6 | 0 | 0 |
| AvgAdjectiveSen | 0.22 | 0.16 | 0.04 | 0.08 | 0 | 0.78 | 0 | 0 |
| AvgAOEBl_IndPolyFAT.3 | 0.21 | 0.26 | 0.05 | 0.29 | 0.04 | 0.24 | 0 | 0.44 |
| AvgAOEBl_InvLinRegSlo | 0.21 | 0.44 | 0.17 | 0.25 | 0 | 0.43 | 0 | 0 |
| AvgDepsBl_dep | 0.21 | 0.09 | 0.17 | 0.01 | 0.69 | 0.57 | 0 | 0 |
| AvgDepsSen_xcomp | 0.2 | 0.26 | 0 | 0.39 | 0 | 0.4 | 0 | 0.29 |
| AvgDepsSen_cop | 0.19 | 0.26 | 0.13 | 0.36 | 0 | 0.31 | 0 | 0.15 |
| AvgDepsBl_compound | 0.18 | 0.07 | 0.17 | 0.01 | 0.45 | 0.23 | 0 | 0.29 |
| LangRhythmDiameter | 0.16 | 0.18 | 0.29 | 0.03 | 0.86 | 0.14 | 0 | 0 |
| AvgConnSen_temp_cons | 0.13 | 0.3 | 0.18 | 0.42 | 0 | 0.09 | 0 | 0 |
| AvgAOEBl_IndAbThr.0.3. | 0.12 | 0.31 | 0.01 | 0.33 | 0.04 | 0.27 | 0 | 0 |
| AvgDepsSen_acl | 0.12 | 0.04 | 0.13 | 0.01 | 0 | 0.3 | 0 | 0 |
| AvgConnSen_order | 0.12 | 0.21 | 0.17 | 0.01 | 0 | 0.23 | 0 | 0 |
| LangRhythmCoeff | 0.12 | 0.41 | 0 | 0.23 | 0 | 0.31 | 0 | 0 |
| AvgSenBlCoh_LeackChod | 0.12 | 0.06 | 0.08 | 0.42 | 0 | 0.28 | 0 | 0 |
| AvgUnqWdBl | 0.11 | 0 | 0 | 1.79 | 0 | 0 | 0 | 0 |
| AvgBlLen | 0.11 | 0 | 0 | 1.8 | 0 | 0 | 0 | 0 |
| AvgVerbBl | 0.1 | 0 | 0 | 1.68 | 0 | 0 | 0 | 0 |
| AvgDepsSen_neg | 0.1 | 0.05 | 0.02 | 0 | 0.08 | 0.37 | 0 | 0 |
| Words | 0.1 | 0 | 0 | 1.75 | 0 | 0 | 0 | 0 |
| Content.words | 0.1 | 0 | 0 | 1.75 | 0 | 0 | 0 | 0 |
| AvgWdBl | 0.1 | 0 | 0 | 1.75 | 0 | 0 | 0 | 0 |
| AvgDepsBl_case | 0.08 | 0 | 0 | 1.27 | 0 | 0 | 0 | 0 |
| AvgNounBl | 0.07 | 0 | 0 | 1.15 | 0 | 0 | 0 | 0 |
| LangRhythmId | 0.07 | 0.02 | 0.23 | 0.02 | 0 | 0 | 0 | 0 |
| AvgPrepositionBl | 0.07 | 0 | 0 | 1.21 | 0 | 0 | 0 | 0 |
| AvgAdverbBl | 0.05 | 0 | 0 | 0.87 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_LDA | 0.04 | 0 | 0 | 0.61 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Shock | 0.04 | 0 | 0 | 0.63 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_Path | 0.04 | 0 | 0 | 0.66 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_word2vec | 0.04 | 0 | 0 | 0.67 | 0 | 0 | 0 | 0 |
| SynSoph | 0.04 | 0 | 0 | 0.68 | 0 | 0 | 0 | 0 |
| Sentences | 0.04 | 0 | 0 | 0.68 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_LSA | 0.04 | 0 | 0 | 0.68 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_word2vec | 0.04 | 0 | 0 | 0.7 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_LSA | 0.03 | 0 | 0 | 0.42 | 0 | 0 | 0 | 0 |
| AvgAOESen_InfPointPoly | 0.03 | 0 | 0 | 0.42 | 0 | 0 | 0 | 0 |
| AvgUnqWdSen | 0.03 | 0 | 0 | 0.43 | 0 | 0 | 0 | 0 |
| AvgConnSen_reas_purp | 0.03 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_LeackChod | 0.03 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgPrepositionSen | 0.03 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgVerbSen | 0.03 | 0 | 0 | 0.46 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_WuPalmer | 0.03 | 0 | 0 | 0.46 | 0 | 0 | 0 | 0 |
| SenStDevUnqWd | 0.03 | 0 | 0 | 0.46 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_WuPalmer | 0.03 | 0 | 0 | 0.46 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Kuperman | 0.03 | 0 | 0 | 0.48 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LDA | 0.03 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0 |
| SenScoreStDev | 0.03 | 0 | 0 | 0.51 | 0 | 0 | 0 | 0 |
| AvgDepsSen_advmod | 0.03 | 0 | 0 | 0.52 | 0 | 0 | 0 | 0 |
| AvgUnqNmdEntBl | 0.03 | 0 | 0 | 0.52 | 0 | 0 | 0 | 0 |
| AvgAdjectiveBl | 0.03 | 0 | 0 | 0.52 | 0 | 0 | 0 | 0 |
| RdbltyFog | 0.03 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Bird | 0.03 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 |
| AvgConnBl_sentence_link | 0.03 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LSA | 0.03 | 0 | 0 | 0.54 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_word2vec | 0.03 | 0 | 0 | 0.55 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_Path | 0.03 | 0 | 0 | 0.55 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Cortese | 0.03 | 0 | 0 | 0.59 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InvLinRegSlo | 0.02 | 0 | 0 | 0.25 | 0 | 0 | 0 | 0 |
| AvgConnSen_sentence_link | 0.02 | 0 | 0 | 0.26 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_IndPolyFAT.3 | 0.02 | 0 | 0 | 0.29 | 0 | 0 | 0 | 0 |
| AvgDepsSen_conj | 0.02 | 0 | 0 | 0.29 | 0 | 0 | 0 | 0 |
| AvgConnBl_coord_conjs | 0.02 | 0 | 0 | 0.3 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InvAverage | 0.02 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 |
| AvgAOEBl_InvAverage | 0.02 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 |
| AvgSenSyll | 0.02 | 0 | 0 | 0.32 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_IndAbThr.0.3. | 0.02 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgDepsBl_auxpass | 0.02 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgConnBl_coord_conns | 0.02 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgSemDep | 0.02 | 0 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| AvgSenStressedSyll | 0.02 | 0 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| AvgSenLen | 0.02 | 0 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InfPointPoly | 0.02 | 0 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| AvgAOEBl_InfPointPoly | 0.02 | 0 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| AvgAOESen_IndAbThr.0.3. | 0.02 | 0 | 0 | 0.35 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_LDA | 0.02 | 0 | 0 | 0.35 | 0 | 0 | 0 | 0 |
| AvgDepsSen_nsubj | 0.02 | 0 | 0 | 0.35 | 0 | 0 | 0 | 0 |
| AvgAOESen_IndPolyFAT.3 | 0.02 | 0 | 0 | 0.36 | 0 | 0 | 0 | 0 |
| AvgWdSen | 0.02 | 0 | 0 | 0.37 | 0 | 0 | 0 | 0 |
| AvgVoice | 0.02 | 0 | 0 | 0.37 | 0 | 0 | 0 | 0 |
| WdMaxDpthHypernymTree | 0.02 | 0 | 0 | 0.38 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LeackChod | 0.02 | 0 | 0 | 0.38 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_WuPalmer | 0.02 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 |
| AvgAOESen_InvAverage | 0.02 | 0 | 0 | 0.41 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Bristol | 0.02 | 0 | 0 | 0.41 | 0 | 0 | 0 | 0 |
| RdbltyKincaid | 0.02 | 0 | 0 | 0.41 | 0 | 0 | 0 | 0 |
| AvgDepsSen_case | 0.02 | 0 | 0 | 0.42 | 0 | 0 | 0 | 0 |
| AvgDepsBl_conj | 0.01 | 0 | 0 | 0.11 | 0 | 0 | 0 | 0 |
| AvgConnSen_coord_conns | 0.01 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 |
| AvgConnSen_conjunctions | 0.01 | 0 | 0 | 0.15 | 0 | 0 | 0 | 0 |
| AvgConnBl_conjunctions | 0.01 | 0 | 0 | 0.15 | 0 | 0 | 0 | 0 |
| AvgDepsSen_cc | 0.01 | 0 | 0 | 0.17 | 0 | 0 | 0 | 0 |
| AvgDepsBl_cc | 0.01 | 0 | 0 | 0.18 | 0 | 0 | 0 | 0 |
| AvgRhythmUnitSyll | 0.01 | 0 | 0 | 0.18 | 0 | 0 | 0 | 0 |
| AvgConnSen_logical_cons | 0.01 | 0 | 0 | 0.18 | 0 | 0 | 0 | 0 |
| AvgConnSen_contrasts | 0 | 0 | 0 | 0.05 | 0 | 0 | 0 | 0 |
| AvgConnSen_coord_conjs | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0 |
ReaderBench Model 1b
This model was trained on winter data in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -2.0039 | 0.3112 | 0.1353 | 0.2667 | -0.0102 | 0.1234 | 0.0268 | 0.222 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| WdEnt | 11.34 | 23.56 | 2.43 | 1.65 | 0.74 | 3.11 | 31.83 | 12.9 |
| AvgDepsBl_det | 6.98 | 12.06 | 2.13 | 1.17 | 2.37 | 2.1 | 9.48 | 12.41 |
| AvgPronounBl | 3.71 | 2.92 | 1.9 | 0.99 | 0 | 1.27 | 7.58 | 10.22 |
| AvgUnqVerbBl | 3.43 | 6.53 | 2.29 | 1.36 | 1.89 | 2.17 | 0 | 3.65 |
| AvgDepsBl_nsubj | 2.97 | 5.88 | 2.3 | 1.43 | 1.14 | 2.11 | 0 | 2.19 |
| AvgUnqPrepositionBl | 2.78 | 4.75 | 2.01 | 1.04 | 1.59 | 1.86 | 0 | 3.65 |
| LxcDiv | 2.6 | 4 | 2.33 | 1.44 | 0.22 | 1.92 | 0 | 3.16 |
| WdDiffWdStem | 1.99 | 0.84 | 0.97 | 0.5 | 0.82 | 0.63 | 0 | 7.3 |
| AvgBlScore | 1.56 | 2.98 | 1.74 | 1.22 | 1.18 | 1.81 | 0 | 0 |
| AvgDepsBl_punct | 1.52 | 2.67 | 1.81 | 0.85 | 0.54 | 1.23 | 0 | 0.97 |
| AvgUnqNoundBl | 1.48 | 0.02 | 1.81 | 0.84 | 0.31 | 1.09 | 11.62 | 2.68 |
| AggPronSen_third_person | 1.37 | 0.36 | 0.55 | 0.21 | 0.28 | 0.67 | 18.65 | 2.19 |
| TCorefChainDoc | 1.3 | 0.78 | 1.88 | 0.91 | 0.67 | 1.7 | 0 | 2.19 |
| RdbltyFlesch | 1.3 | 2.18 | 0.2 | 0.44 | 1.65 | 0.98 | 0 | 2.19 |
| AvgPronBl_first_person | 1.19 | 0 | 1.69 | 0.74 | 1.34 | 0.66 | 0 | 3.65 |
| AvgSenBl | 1.16 | 2.23 | 1.8 | 0.84 | 0.27 | 0.97 | 0 | 0 |
| AvgDepsSen_advcl | 1.14 | 0.06 | 0.01 | 0.49 | 0.72 | 0.76 | 0 | 4.62 |
| AvgSenBlCoh_LeackChod | 1.12 | 1.42 | 1.49 | 0.61 | 0.42 | 1.35 | 0 | 1.22 |
| AvgSenBlCoh_LDA | 1.07 | 0.19 | 0.94 | 0.65 | 1.67 | 0.94 | 15.45 | 0.49 |
| LexChainMaxSp | 0.99 | 1.34 | 1.69 | 0.74 | 2.27 | 1.78 | 0 | 0 |
| AvgDepsBl_mark | 0.94 | 0.2 | 1.21 | 0.38 | 1.95 | 0.86 | 0 | 2.68 |
| CharEnt | 0.86 | 0.48 | 1.61 | 0.8 | 0.84 | 0.82 | 0 | 1.22 |
| AvgWdLen | 0.85 | 0.9 | 1.12 | 0.76 | 0.69 | 0.73 | 0 | 0.97 |
| AvgChainSpan | 0.79 | 0.68 | 1.73 | 1.07 | 1.32 | 1.08 | 0 | 0 |
| AvgAOESen_InfPointPoly | 0.73 | 0.22 | 0.35 | 0.23 | 0.12 | 0.39 | 0 | 2.68 |
| AvgDepsSen_det | 0.72 | 0.92 | 0.1 | 0.35 | 0.83 | 0.57 | 0 | 1.46 |
| AvgDepsSen_dobj | 0.69 | 0.6 | 0.53 | 0.4 | 0.94 | 0.52 | 0 | 1.46 |
| AvgUnqPronounBl | 0.66 | 0.14 | 1.72 | 0.76 | 0.12 | 1.13 | 0 | 0.49 |
| AvgConnBl_temp_conns | 0.65 | 1.22 | 1.12 | 0.33 | 1.18 | 0.69 | 0 | 0 |
| AvgDepsSen_compound | 0.64 | 0.99 | 0.55 | 0.18 | 1.46 | 1.18 | 0 | 0.49 |
| AvgSenAdjCoh_Path | 0.64 | 0.78 | 1.29 | 0.71 | 0.77 | 0.79 | 0 | 0 |
| WdDiffLemmaStem | 0.63 | 1.33 | 0.19 | 0.58 | 0.14 | 0.74 | 0 | 0 |
| RdbltyDaleChall | 0.63 | 1.19 | 1.14 | 0.4 | 0.66 | 0.48 | 0 | 0 |
| WdMaxDpthHypernymTree | 0.59 | 1.07 | 0.69 | 0.6 | 0.39 | 0.51 | 0 | 0 |
| LexChainAvgSpan | 0.57 | 0.28 | 0.88 | 0.7 | 1.27 | 0.59 | 3.91 | 0 |
| SenStdDevWd | 0.54 | 0.55 | 0.97 | 0.56 | 0.03 | 0.68 | 0 | 0.24 |
| AvgDepsSen_amod | 0.54 | 0.18 | 0.25 | 0.34 | 0.58 | 0.73 | 0 | 1.46 |
| AvgDepsBl_dobj | 0.54 | 0.02 | 1.67 | 0.72 | 2.01 | 0.84 | 0 | 0.24 |
| AvgDepsSen_dep | 0.52 | 0.87 | 0.47 | 0.4 | 0.18 | 1.08 | 0 | 0 |
| AvgDepsBl_nmod | 0.52 | 0.05 | 1.74 | 0.78 | 0.19 | 0.95 | 0 | 0 |
| FrqRhythmId | 0.51 | 0.56 | 1.17 | 0.42 | 0.91 | 0.9 | 0 | 0 |
| AvgDepsBl_advcl | 0.5 | 0.1 | 1.08 | 0.3 | 1.09 | 0.56 | 0 | 0.97 |
| AvgCorefChain | 0.5 | 0.3 | 1.13 | 0.49 | 1.28 | 0.45 | 0 | 0.49 |
| AvgDepsSen_nmod | 0.49 | 0.06 | 0.34 | 0.5 | 0.22 | 0.58 | 0 | 1.22 |
| AvgConnSen_addition | 0.49 | 0.1 | 0.62 | 0.49 | 0.44 | 0.68 | 0 | 0.97 |
| AvgAdjectiveBl | 0.48 | 0.23 | 1.51 | 0.59 | 0.56 | 0.7 | 0 | 0 |
| WdLettStdDev | 0.46 | 0.17 | 1.22 | 0.74 | 0.3 | 0.72 | 0 | 0 |
| AvgPronBl_indefinite | 0.44 | 0.23 | 1.25 | 0.4 | 0.87 | 1.02 | 0 | 0 |
| AvgConnBl_addition | 0.44 | 0.63 | 0.99 | 0.25 | 0.74 | 0.66 | 0 | 0 |
| AvgDepsSen_mark | 0.44 | 0.16 | 0.1 | 0.39 | 1.46 | 0.73 | 0 | 0.97 |
| LangRhythmCoeff | 0.44 | 0.71 | 0.54 | 0.34 | 0.04 | 0.82 | 0 | 0 |
| AvgVoice | 0.43 | 0.26 | 1.31 | 0.44 | 0.83 | 0.69 | 0 | 0 |
| AvgUnqAdverbBl | 0.43 | 0.03 | 1.52 | 0.6 | 0.67 | 0.76 | 0 | 0 |
| AvgAOABl_Shock | 0.43 | 0.25 | 1.21 | 0.44 | 0.69 | 0.88 | 0 | 0 |
| AvgBlLen | 0.41 | 0 | 0 | 1.69 | 0 | 0 | 0 | 0 |
| AvgPronBl_third_person | 0.41 | 0.2 | 1.12 | 0.32 | 0.64 | 0.33 | 0 | 0.49 |
| AvgUnqWdBl | 0.41 | 0 | 0 | 1.7 | 0 | 0 | 0 | 0 |
| Content.words | 0.4 | 0 | 0 | 1.67 | 0 | 0 | 0 | 0 |
| AvgWdBl | 0.4 | 0 | 0 | 1.67 | 0 | 0 | 0 | 0 |
| Words | 0.39 | 0 | 0 | 1.6 | 0 | 0 | 0 | 0 |
| AvgAOEBl_InfPointPoly | 0.39 | 0.14 | 0.34 | 0.24 | 0.81 | 0.48 | 0 | 0.97 |
| TCorefChainBigSpan | 0.38 | 0 | 1.06 | 0.29 | 1.22 | 1.08 | 1.47 | 0 |
| AvgDepsBl_cop | 0.37 | 0 | 1.4 | 0.51 | 1.32 | 0.58 | 0 | 0 |
| AvgConnSen_semi_coords | 0.37 | 0.06 | 0.12 | 0.29 | 0.84 | 0.63 | 0 | 0.97 |
| AvgVerbBl | 0.37 | 0 | 0 | 1.55 | 0 | 0 | 0 | 0 |
| AvgDepsSen_xcomp | 0.35 | 0.38 | 0.05 | 0.6 | 1.2 | 0.67 | 0 | 0 |
| AvgRhythmUnitStreesSyll | 0.34 | 0.4 | 0.2 | 0.33 | 1.14 | 0.93 | 0 | 0 |
| AvgAOEBl_InvLinRegSlo | 0.34 | 0.7 | 0.09 | 0.23 | 0.67 | 0.59 | 0 | 0 |
| AvgNmdEntSen | 0.34 | 0.3 | 0.21 | 0.4 | 0.48 | 1.1 | 0 | 0 |
| AvgConnBl_coord_connects | 0.34 | 0 | 1.25 | 0.4 | 1.85 | 0.63 | 0 | 0 |
| AvgAOABl_Bird | 0.34 | 0.25 | 0.44 | 0.41 | 1.53 | 0.92 | 0 | 0 |
| AvgDepsBl_xcomp | 0.33 | 0.18 | 1.15 | 0.34 | 0 | 0.45 | 0 | 0 |
| AvgAdverbSen | 0.33 | 0.27 | 0.2 | 0.4 | 2.52 | 0.97 | 0 | 0 |
| AvgConnBl_reas_purp | 0.33 | 0 | 0.96 | 0.24 | 1.1 | 0.45 | 0 | 0.49 |
| AvgAdverbBl | 0.33 | 0.04 | 1.29 | 0.43 | 0.75 | 0.5 | 0 | 0 |
| TActCorefChainWd | 0.33 | 0.62 | 0.39 | 0.29 | 0.22 | 0.34 | 0 | 0 |
| AvgAOABl_Bristol | 0.32 | 0.11 | 0.75 | 0.33 | 0.23 | 1 | 0 | 0 |
| AvgAOEBl_IndexPolyFAT.3 | 0.32 | 0.58 | 0 | 0.44 | 0.44 | 0.43 | 0 | 0 |
| AvgDepsBl_aux | 0.32 | 0 | 0.52 | 0.07 | 0.8 | 0.39 | 0 | 0.97 |
| AvgConnSen_logical_conns | 0.31 | 0.13 | 0.7 | 0.47 | 0.7 | 0.55 | 0 | 0 |
| AvgDepsSen_punct | 0.31 | 0.05 | 0.9 | 0.55 | 0.75 | 0.45 | 0 | 0 |
| AvgCommaBl | 0.3 | 0 | 1.16 | 0.35 | 0.23 | 0.69 | 0 | 0 |
| AvgDepsSen_aux | 0.29 | 0.02 | 0.45 | 0.36 | 0.43 | 1.18 | 0 | 0 |
| AvgNounBl | 0.29 | 0 | 0 | 1.2 | 0 | 0 | 0 | 0 |
| WdPolysemyCnt | 0.29 | 0.56 | 0.15 | 0.14 | 0.03 | 0.68 | 0 | 0 |
| AvgConnSen_reas_purp | 0.29 | 0.02 | 0.16 | 0.26 | 1.13 | 0.85 | 0 | 0.49 |
| AvgDepsBl_compound | 0.29 | 0.02 | 0.12 | 0 | 1.6 | 0.57 | 0 | 0.97 |
| AvgDepsBl_amod | 0.28 | 0 | 1.1 | 0.32 | 0.78 | 0.53 | 0 | 0 |
| AvgAOASen_Bird | 0.28 | 0.51 | 0.15 | 0.23 | 1.13 | 0.46 | 0 | 0 |
| WdPathCntHypernymTree | 0.28 | 0.06 | 0.7 | 0.51 | 0.06 | 0.52 | 0 | 0 |
| AvgDepsBl_acl | 0.27 | 0.03 | 1 | 0.26 | 0.7 | 0.59 | 0 | 0 |
| AvgDepsBl_ccomp | 0.27 | 0 | 0.9 | 0.21 | 0.92 | 0.86 | 0 | 0 |
| AvgAOASen_Shock | 0.27 | 0.02 | 0.53 | 0.5 | 0.19 | 0.66 | 0 | 0 |
| AggPronSen_indefinite | 0.27 | 0.07 | 0.04 | 0.61 | 0.75 | 0.79 | 0 | 0 |
| AvgDepsSen_cop | 0.27 | 0.16 | 0.03 | 0.55 | 0.5 | 0.75 | 0 | 0 |
| AvgConnBl_simp_subords | 0.27 | 0.06 | 1.07 | 0.3 | 0.95 | 0.41 | 0 | 0 |
| AvgConnBl_semi_coords | 0.26 | 0 | 0.68 | 0.12 | 0.22 | 0.42 | 0 | 0.49 |
| AvgAOESen_IndexAbThr.0.3. | 0.26 | 0.1 | 0.23 | 0.53 | 0.02 | 0.64 | 0 | 0 |
| AvgAOEBl_IndexAbThr.0.3. | 0.26 | 0.21 | 0 | 0.61 | 0.28 | 0.43 | 0 | 0 |
| WdSylCnt | 0.25 | 0.15 | 0.39 | 0.48 | 0.97 | 0.3 | 0 | 0 |
| AvgDepsBl_neg | 0.25 | 0.03 | 0.86 | 0.19 | 0.09 | 0.84 | 0 | 0 |
| AvgConnBl_contrasts | 0.23 | 0 | 0.73 | 0.14 | 0.65 | 0.85 | 0 | 0 |
| AvgNounSen | 0.23 | 0.05 | 0.42 | 0.24 | 0.23 | 0.89 | 0 | 0 |
| AvgDepsSen_ccomp | 0.23 | 0.18 | 0.07 | 0.29 | 0.47 | 0.83 | 0 | 0 |
| AvgConnBl_logical_conns | 0.23 | 0.16 | 0.87 | 0.19 | 0.05 | 0.27 | 0 | 0 |
| AvgConnSen_simp_subords | 0.23 | 0.1 | 0.05 | 0.54 | 0 | 0.54 | 0 | 0 |
| AvgAdjectiveSen | 0.23 | 0.1 | 0.21 | 0.34 | 0.85 | 0.79 | 0 | 0 |
| AvgConnBl_oppositions | 0.23 | 0 | 0.79 | 0.16 | 0.75 | 0.8 | 0 | 0 |
| AvgInferenceDistChain | 0.23 | 0.1 | 0.42 | 0.26 | 0.53 | 0.76 | 0 | 0 |
| AvgConnBl_order | 0.22 | 0 | 0.9 | 0.21 | 2.32 | 0.29 | 0 | 0 |
| AvgDepsBl_conj | 0.22 | 0 | 0.79 | 0.16 | 1.65 | 0.57 | 0 | 0 |
| AvgPrepositionBl | 0.22 | 0 | 0 | 0.9 | 0 | 0 | 0 | 0 |
| AvgSenScore | 0.22 | 0.05 | 0.14 | 0.33 | 0.33 | 0.9 | 0 | 0 |
| AvgDepsBl_case | 0.22 | 0 | 0 | 0.9 | 0 | 0 | 0 | 0 |
| AvgAOESen_IndexPolyFAT.3 | 0.22 | 0.07 | 0.26 | 0.42 | 0.35 | 0.55 | 0 | 0 |
| AvgAOABl_Cortese | 0.22 | 0.05 | 0.35 | 0.32 | 1.05 | 0.67 | 0 | 0 |
| AvgConnSen_oppositions | 0.21 | 0.03 | 0.03 | 0.35 | 1.28 | 0.84 | 0 | 0 |
| AvgIntraBlCoh_word2vec | 0.21 | 0 | 0 | 0.88 | 0 | 0 | 0 | 0 |
| AvgRhythmUnits | 0.21 | 0 | 0.34 | 0.42 | 1.08 | 0.53 | 0 | 0 |
| AvgNounNmdEntBl | 0.2 | 0.14 | 0.52 | 0.07 | 2.7 | 0.46 | 0 | 0 |
| Sentences | 0.2 | 0 | 0 | 0.84 | 0 | 0 | 0 | 0 |
| AvgAOABl_Kuperman | 0.2 | 0.13 | 0.06 | 0.32 | 0.93 | 0.64 | 0 | 0 |
| AvgIntraBlCoh_LDA | 0.2 | 0 | 0 | 0.84 | 0 | 0 | 0 | 0 |
| AvgNmdEntBl | 0.2 | 0.03 | 0.82 | 0.18 | 1.51 | 0.35 | 0 | 0 |
| AvgConnSen_order | 0.19 | 0.11 | 0.13 | 0.37 | 0.55 | 0.41 | 0 | 0 |
| AvgConnSen_temp_conns | 0.19 | 0.27 | 0.12 | 0 | 1.47 | 0.77 | 0 | 0 |
| LxcSoph | 0.19 | 0.05 | 0.18 | 0.34 | 0.45 | 0.61 | 0 | 0 |
| LangRhythmDiameter | 0.19 | 0 | 0.3 | 0.02 | 1.45 | 0.37 | 0 | 0.49 |
| AvgIntraBlCoh_LSA | 0.19 | 0 | 0 | 0.81 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_Path | 0.19 | 0 | 0 | 0.81 | 0 | 0 | 0 | 0 |
| AvgAOASen_Kuperman | 0.18 | 0.28 | 0.06 | 0.17 | 0.06 | 0.45 | 0 | 0 |
| AvgDepsBl_nsubjpass | 0.18 | 0.04 | 0.66 | 0.11 | 0.38 | 0.53 | 0 | 0 |
| AvgAOASen_Bristol | 0.18 | 0.13 | 0.18 | 0.25 | 0.36 | 0.55 | 0 | 0 |
| AvgSenAdjCoh_LDA | 0.17 | 0 | 0 | 0.69 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_Path | 0.17 | 0 | 0 | 0.69 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LeackChod | 0.17 | 0 | 0 | 0.7 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_LeackChod | 0.17 | 0 | 0 | 0.71 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_word2vec | 0.17 | 0 | 0 | 0.71 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_WuPalmer | 0.17 | 0 | 0 | 0.72 | 0 | 0 | 0 | 0 |
| SenAsson | 0.16 | 0 | 0.47 | 0.06 | 0.6 | 0.7 | 0 | 0 |
| AvgDepsBl_nummod | 0.16 | 0 | 0.47 | 0.06 | 0.07 | 0.75 | 0 | 0 |
| AvgSenBlCoh_word2vec | 0.16 | 0 | 0 | 0.66 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_WuPalmer | 0.16 | 0 | 0 | 0.66 | 0 | 0 | 0 | 0 |
| SenStDevUnqWd | 0.15 | 0 | 0 | 0.61 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_IndexAbThr.0.3. | 0.15 | 0 | 0 | 0.61 | 0 | 0 | 0 | 0 |
| AvgUnqWdSen | 0.15 | 0 | 0 | 0.62 | 0 | 0 | 0 | 0 |
| SynSoph | 0.15 | 0 | 0 | 0.63 | 0 | 0 | 0 | 0 |
| AvgUnqAdjectiveBl | 0.15 | 0 | 0 | 0.63 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LSA | 0.15 | 0 | 0 | 0.63 | 0 | 0 | 0 | 0 |
| AvgWdSen | 0.14 | 0 | 0 | 0.56 | 0 | 0 | 0 | 0 |
| AvgSenLen | 0.14 | 0 | 0 | 0.56 | 0 | 0 | 0 | 0 |
| AvgConnBl_sentence_link | 0.14 | 0 | 0 | 0.57 | 0 | 0 | 0 | 0 |
| WdAvgDpthHypernymTree | 0.14 | 0 | 0 | 0.58 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_LSA | 0.14 | 0 | 0 | 0.58 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_WuPalmer | 0.14 | 0 | 0 | 0.6 | 0 | 0 | 0 | 0 |
| AvgDepsSen_mwe | 0.13 | 0 | 0.31 | 0.03 | 0.32 | 0.7 | 0 | 0 |
| AvgDepsBl_advmod | 0.13 | 0 | 0 | 0.54 | 0 | 0 | 0 | 0 |
| AvgDepsSen_advmod | 0.13 | 0 | 0 | 0.54 | 0 | 0 | 0 | 0 |
| AvgConnSen_contrasts | 0.13 | 0 | 0.14 | 0.22 | 0.06 | 0.55 | 0 | 0 |
| AvgDepsSen_nsubj | 0.12 | 0 | 0 | 0.48 | 0 | 0 | 0 | 0 |
| SenScoreStDev | 0.12 | 0 | 0 | 0.49 | 0 | 0 | 0 | 0 |
| AvgVerbSen | 0.12 | 0 | 0 | 0.49 | 0 | 0 | 0 | 0 |
| AvgSenStressedSyll | 0.12 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0 |
| AvgBlVoiceCoOcc | 0.12 | 0 | 0 | 0.51 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Shock | 0.11 | 0 | 0 | 0.44 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_IndexPolyFAT.3 | 0.11 | 0 | 0 | 0.44 | 0 | 0 | 0 | 0 |
| AvgSemDep | 0.11 | 0 | 0 | 0.44 | 0 | 0 | 0 | 0 |
| AvgDepsSen_case | 0.11 | 0 | 0 | 0.46 | 0 | 0 | 0 | 0 |
| AvgRhythmUnitSyll | 0.1 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 |
| AvgSenSyll | 0.1 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Bird | 0.1 | 0 | 0 | 0.41 | 0 | 0 | 0 | 0 |
| AvgDepsSen_acl | 0.1 | 0.06 | 0.2 | 0.01 | 0.6 | 0.43 | 0 | 0 |
| RdbltyKincaid | 0.1 | 0 | 0 | 0.42 | 0 | 0 | 0 | 0 |
| AvgAOASen_Cortese | 0.1 | 0.12 | 0.06 | 0.25 | 0.22 | 0 | 0 | 0 |
| AvgConnBl_conjunctions | 0.09 | 0 | 0 | 0.36 | 0 | 0 | 0 | 0 |
| AvgPronounSen | 0.09 | 0 | 0 | 0.37 | 0 | 0 | 0 | 0 |
| RdbltyFog | 0.09 | 0 | 0 | 0.37 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Cortese | 0.08 | 0 | 0 | 0.32 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Kuperman | 0.08 | 0 | 0 | 0.32 | 0 | 0 | 0 | 0 |
| AvgPrepositionSen | 0.08 | 0 | 0 | 0.32 | 0 | 0 | 0 | 0 |
| AvgDepsBl_dep | 0.08 | 0.04 | 0.25 | 0.02 | 0.15 | 0.29 | 0 | 0 |
| AvgDepsBl_cc | 0.08 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Bristol | 0.08 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgConnSen_sentence_link | 0.08 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgDepsSen_neg | 0.08 | 0 | 0.13 | 0 | 0.14 | 0.58 | 0 | 0 |
| AvgDepsSen_conj | 0.08 | 0 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| AvgDepsBl_mwe | 0.08 | 0 | 0.43 | 0.05 | 0.03 | 0.18 | 0 | 0 |
| AvgConnSen_coord_connects | 0.07 | 0 | 0 | 0.29 | 0 | 0 | 0 | 0 |
| AvgConnSen_coord_conjs | 0.07 | 0 | 0 | 0.29 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InvAverage | 0.07 | 0 | 0 | 0.29 | 0 | 0 | 0 | 0 |
| AvgAOEBl_InvAverage | 0.07 | 0 | 0 | 0.29 | 0 | 0 | 0 | 0 |
| AvgAOESen_InvAverage | 0.07 | 0 | 0 | 0.3 | 0 | 0 | 0 | 0 |
| AvgDepsSen_cc | 0.07 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 |
| AvgConnSen_conjunctions | 0.07 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InfPointPoly | 0.06 | 0 | 0 | 0.24 | 0 | 0 | 0 | 0 |
| AvgUnqNmdEntBl | 0.06 | 0 | 0 | 0.24 | 0 | 0 | 0 | 0 |
| LangRhythmId | 0.06 | 0.03 | 0.04 | 0 | 0.33 | 0.37 | 0 | 0 |
| AvgAOESen_InvLinRegSlo | 0.05 | 0 | 0 | 0.22 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InvLinRegSlo | 0.05 | 0 | 0 | 0.23 | 0 | 0 | 0 | 0 |
| AvgDepsBl_auxpass | 0.04 | 0 | 0 | 0.18 | 0 | 0 | 0 | 0 |
| AvgConnBl_coord_conjs | 0.03 | 0 | 0 | 0.12 | 0 | 0 | 0 | 0 |
ReaderBench Model 1c
This model was trained on spring data in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -5.6692 | 0.1651 | 0.2625 | 0.1043 | -0.0146 | 0.4555 | 0.1632 | -0.0348 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| WdEnt | 7.76 | 15.63 | 2.27 | 1.53 | 2 | 3.41 | 25.87 | 9.83 |
| AvgUnqVerbBl | 5.89 | 24.09 | 2.26 | 1.36 | 2.3 | 3.86 | 0 | 12.86 |
| AvgBlScore | 2.6 | 7.12 | 1.53 | 1.13 | 0.58 | 2.67 | 0 | 4.69 |
| AvgNounSen | 2.57 | 0.61 | 0.6 | 0.39 | 1.06 | 1.04 | 14.54 | 2.12 |
| LxcDiv | 2.18 | 3.58 | 2.18 | 1.33 | 0.72 | 2.54 | 0 | 3.78 |
| AvgDepsSen_compound | 2.17 | 1.45 | 1.12 | 0.56 | 0.27 | 1.54 | 7.94 | 2.12 |
| WdDiffLemmaStem | 2.07 | 0.57 | 0.93 | 0.49 | 0.22 | 1 | 10.51 | 0 |
| AvgDepsBl_dobj | 2.06 | 0.09 | 1.63 | 0.74 | 0.92 | 1.13 | 9.09 | 0 |
| AvgUnqPrepositionBl | 1.96 | 1.98 | 2.25 | 1.27 | 1.28 | 2.47 | 0 | 5.6 |
| AvgDepsBl_punct | 1.85 | 2.48 | 1.93 | 0.93 | 1.96 | 2.21 | 0 | 6.2 |
| AvgDepsBl_nsubj | 1.84 | 3.1 | 1.97 | 1.26 | 0.88 | 2.06 | 0 | 1.36 |
| RdbltyFlesch | 1.75 | 0.27 | 0.22 | 0.16 | 1.12 | 0.23 | 12.02 | 0.15 |
| AvgWdLen | 1.74 | 3.11 | 1.54 | 0.76 | 0.21 | 2.1 | 0 | 3.03 |
| AvgDepsBl_nmod | 1.65 | 2.75 | 1.94 | 0.92 | 0.42 | 1.77 | 0 | 2.42 |
| AvgUnqNoundBl | 1.28 | 0.06 | 0.99 | 0.68 | 1.79 | 0.1 | 6.94 | 2.72 |
| AvgPronounBl | 1.28 | 0.4 | 1.85 | 1.14 | 0.98 | 1.75 | 0 | 1.21 |
| AvgUnqPronounBl | 1.23 | 0.36 | 1.69 | 0.74 | 0.27 | 0.86 | 3 | 0.76 |
| WdSylCnt | 1.22 | 0.89 | 1.39 | 0.58 | 1.52 | 1.62 | 0 | 5.45 |
| AvgUnqAdjectiveBl | 1.19 | 0.24 | 1.37 | 0.54 | 0.34 | 0.6 | 4.3 | 0.76 |
| AvgDepsBl_ccomp | 1.18 | 0.35 | 0.61 | 0.05 | 0.35 | 0.61 | 5.78 | 0.76 |
| AvgChainSpan | 1.17 | 0.51 | 1.66 | 0.99 | 0.49 | 1.6 | 0 | 0.91 |
| LexChainMaxSp | 1.14 | 0.38 | 1.67 | 0.75 | 1.99 | 1.62 | 0 | 0 |
| WdDiffWdStem | 1.11 | 0.76 | 1.38 | 0.78 | 0.06 | 1.6 | 0 | 0 |
| AvgDepsBl_det | 1.1 | 0.29 | 1.71 | 0.72 | 0.82 | 1.52 | 0 | 0.76 |
| WdLettStdDev | 1.02 | 1.03 | 1.66 | 0.85 | 1.56 | 1.04 | 0 | 0.3 |
| FrqRhythmId | 1 | 0.65 | 1.44 | 0.59 | 0.95 | 1.32 | 0 | 0.76 |
| AvgAOABl_Shock | 0.97 | 0.91 | 1.13 | 0.67 | 0.02 | 1.34 | 0 | 0 |
| AvgSenBlCoh_LDA | 0.97 | 1.01 | 1.13 | 0.82 | 1.53 | 1.24 | 0 | 0 |
| AvgDepsBl_mark | 0.96 | 0.04 | 1.52 | 0.65 | 1.46 | 1.4 | 0 | 0 |
| AvgSenAdjCoh_word2vec | 0.9 | 0.39 | 1.37 | 0.67 | 0.29 | 1.18 | 0 | 1.06 |
| TCorefChainDoc | 0.87 | 0.32 | 1.67 | 0.71 | 2.43 | 0.96 | 0 | 0 |
| AvgDepsSen_punct | 0.81 | 0.4 | 1 | 0.68 | 0.26 | 1.2 | 0 | 0 |
| LangRhythmCoeff | 0.8 | 0.47 | 0.95 | 0.46 | 0.2 | 1.23 | 0 | 0 |
| AvgPronBl_first_person | 0.78 | 0.58 | 1.31 | 0.44 | 0.15 | 0.86 | 0 | 1.66 |
| RdbltyDaleChall | 0.78 | 1.44 | 0.96 | 0.38 | 1.08 | 0.76 | 0 | 1.06 |
| AvgConnBl_sentence_link | 0.74 | 0.09 | 1.31 | 0.45 | 3.5 | 0.92 | 0 | 0.3 |
| AvgDepsBl_compound | 0.74 | 1.08 | 0.77 | 0.12 | 1.07 | 0.86 | 0 | 3.48 |
| AvgConnSen_logical_conns | 0.71 | 0.31 | 0.54 | 0.51 | 0.72 | 1.24 | 0 | 0.76 |
| LexChainAvgSpan | 0.7 | 0.67 | 1.02 | 0.62 | 0.5 | 0.81 | 0 | 0 |
| AvgUnqAdverbBl | 0.69 | 0.01 | 1.46 | 0.57 | 0.78 | 0.75 | 0 | 0.76 |
| CharEnt | 0.66 | 0.48 | 1.35 | 0.78 | 1.51 | 0.49 | 0 | 0.76 |
| AvgDepsBl_amod | 0.66 | 0.23 | 1.31 | 0.48 | 1.2 | 0.73 | 0 | 0 |
| AvgRhythmUnitStreesSyll | 0.65 | 0.39 | 0.43 | 0.21 | 0.86 | 1.22 | 0 | 0 |
| AvgAdjectiveSen | 0.64 | 0.7 | 0.68 | 0.45 | 0.03 | 0.78 | 0 | 2.72 |
| AvgDepsBl_advcl | 0.62 | 0.07 | 1.21 | 0.42 | 1.83 | 0.74 | 0 | 0 |
| AvgConnBl_simp_subords | 0.61 | 0.04 | 1.25 | 0.45 | 0.28 | 0.69 | 0 | 0.76 |
| AvgConnBl_reas_purp | 0.61 | 0.57 | 0.98 | 0.27 | 0.17 | 0.74 | 0 | 0 |
| AvgCorefChain | 0.6 | 0.54 | 1.17 | 0.48 | 1.94 | 0.52 | 0 | 0 |
| AvgPronBl_indefinite | 0.59 | 0.2 | 1.33 | 0.5 | 0.05 | 0.57 | 0 | 0 |
| AvgDepsBl_xcomp | 0.58 | 0.36 | 1.14 | 0.37 | 0.09 | 0.63 | 0 | 0 |
| AvgBlVoiceCoOcc | 0.57 | 0 | 1.44 | 0.56 | 0.17 | 0.51 | 0 | 0 |
| AvgDepsBl_aux | 0.56 | 0.17 | 1.12 | 0.32 | 1.5 | 0.64 | 0 | 0 |
| AvgDepsBl_mwe | 0.55 | 0 | 0.93 | 0.23 | 1.62 | 0.79 | 0 | 0 |
| AvgPronBl_third_person | 0.55 | 0.13 | 1.23 | 0.39 | 0.24 | 0.53 | 0 | 1.06 |
| AggPronSen_third_person | 0.55 | 0.37 | 0.47 | 0.21 | 2.96 | 0.87 | 0 | 0.76 |
| AvgAOABl_Cortese | 0.55 | 0.56 | 0.64 | 0.7 | 0.31 | 0.68 | 0 | 0 |
| AvgDepsBl_neg | 0.54 | 0.08 | 0.71 | 0.15 | 0.29 | 0.91 | 0 | 0 |
| AvgConnSen_order | 0.54 | 0.06 | 0.26 | 0.52 | 0.98 | 1.07 | 0 | 0 |
| AvgDepsSen_amod | 0.53 | 0.43 | 0.6 | 0.49 | 0.07 | 0.72 | 0 | 0.61 |
| AvgInferenceDistChain | 0.53 | 0.52 | 0.46 | 0.31 | 1.67 | 0.81 | 0 | 0 |
| TCorefChainBigSpan | 0.52 | 0.04 | 1.24 | 0.35 | 0.13 | 0.53 | 0 | 0 |
| SenAsson | 0.51 | 0.33 | 0.85 | 0.21 | 1.05 | 0.63 | 0 | 0.15 |
| AvgConnBl_contrasts | 0.5 | 0.06 | 0.85 | 0.21 | 0 | 0.72 | 0 | 0 |
| AggPronSen_indefinite | 0.49 | 0.31 | 0.15 | 0.39 | 0.83 | 0.93 | 0 | 0.76 |
| AvgDepsSen_xcomp | 0.48 | 0.76 | 0.22 | 0.37 | 0.33 | 0.72 | 0 | 0 |
| AvgConnBl_temp_conns | 0.47 | 0.2 | 1.04 | 0.26 | 0.13 | 0.46 | 0 | 0.61 |
| AvgDepsBl_cop | 0.46 | 0.25 | 0.92 | 0.23 | 0.13 | 0.5 | 0 | 0 |
| AvgAOASen_Kuperman | 0.46 | 0.19 | 0.45 | 0.37 | 1.7 | 0.71 | 0 | 0.61 |
| AvgDepsSen_nmod | 0.46 | 0.2 | 0.11 | 0.46 | 1.37 | 0.74 | 0 | 4.39 |
| AvgDepsSen_dobj | 0.45 | 0.17 | 0.44 | 0.42 | 0.55 | 0.74 | 0 | 0 |
| AvgDepsBl_nummod | 0.44 | 0.07 | 0.35 | 0.02 | 0.46 | 0.88 | 0 | 0 |
| AvgAOEBl_IndexPolyFAT.3 | 0.44 | 0.19 | 0.42 | 0.4 | 1.29 | 0.71 | 0 | 0 |
| AvgConnBl_order | 0.42 | 0.05 | 0.62 | 0.11 | 0.53 | 0.67 | 0 | 0 |
| LxcSoph | 0.42 | 0.28 | 0.06 | 0.07 | 0.93 | 0.88 | 0 | 0.76 |
| AvgDepsSen_neg | 0.42 | 0.06 | 0.25 | 0.02 | 0.84 | 0.91 | 0 | 0 |
| AvgConnSen_simp_subords | 0.41 | 0.36 | 0.07 | 0.56 | 0.46 | 0.74 | 0 | 0 |
| SenStdDevWd | 0.4 | 0.08 | 0.88 | 0.66 | 0.4 | 0.32 | 0 | 0 |
| AvgConnBl_oppositions | 0.4 | 0.02 | 0.95 | 0.24 | 1.8 | 0.37 | 0 | 0 |
| AvgDepsSen_ccomp | 0.4 | 0.1 | 0.57 | 0.44 | 0.58 | 0.47 | 0 | 2.12 |
| AvgAOABl_Kuperman | 0.39 | 0.38 | 0.37 | 0.53 | 0.22 | 0.51 | 0 | 0 |
| AvgRhythmUnits | 0.39 | 0.28 | 0.16 | 0.48 | 0.22 | 0.68 | 0 | 0 |
| AvgAOEBl_InvLinRegSlo | 0.37 | 0.13 | 0.67 | 0.33 | 0.23 | 0.41 | 0 | 0.76 |
| TActCorefChainWd | 0.37 | 0.29 | 0.51 | 0.26 | 0.8 | 0.49 | 0 | 0 |
| AvgPronounSen | 0.36 | 0.32 | 0.6 | 0.37 | 0.49 | 0.33 | 0 | 0.76 |
| AvgDepsSen_mwe | 0.36 | 0.01 | 0.33 | 0.03 | 0.68 | 0.73 | 0 | 0 |
| AvgAOASen_Bristol | 0.35 | 0.6 | 0.24 | 0.13 | 0.88 | 0.42 | 0 | 1.36 |
| LangRhythmDiameter | 0.34 | 0.13 | 0.23 | 0.01 | 0.31 | 0.68 | 0 | 0 |
| AvgCommaBl | 0.34 | 0.07 | 0.66 | 0.12 | 0.49 | 0.43 | 0 | 0 |
| AvgAOASen_Cortese | 0.34 | 0.33 | 0.69 | 0.45 | 0.27 | 0.24 | 0 | 0 |
| AvgDepsSen_mark | 0.34 | 0.11 | 0.22 | 0.54 | 0.34 | 0.58 | 0 | 0 |
| AvgDepsSen_acl | 0.33 | 0.04 | 0.71 | 0.13 | 0.93 | 0.38 | 0 | 0 |
| AvgConnBl_logical_conns | 0.33 | 0.05 | 0.53 | 0.06 | 0.26 | 0.51 | 0 | 0 |
| WdPathCntHypernymTree | 0.32 | 0.52 | 0.45 | 0.13 | 0.56 | 0.32 | 0 | 0 |
| AvgConnBl_semi_coords | 0.32 | 0.14 | 0.78 | 0.16 | 0.43 | 0.27 | 0 | 0 |
| AvgDepsSen_cop | 0.31 | 0.2 | 0.47 | 0.47 | 0.17 | 0.34 | 0 | 0 |
| AvgAOESen_InfPointPoly | 0.3 | 0.34 | 0.31 | 0.16 | 1.12 | 0.4 | 0 | 0 |
| AvgConnSen_semi_coords | 0.3 | 0.06 | 0.01 | 0.34 | 0.68 | 0.64 | 0 | 0 |
| AvgAOASen_Shock | 0.3 | 0.28 | 0.21 | 0.49 | 0.06 | 0.4 | 0 | 0.61 |
| AvgConnSen_oppositions | 0.3 | 0.08 | 0.02 | 0 | 0.39 | 0.73 | 0 | 0 |
| AvgDepsSen_advmod | 0.29 | 0.32 | 0.18 | 0.47 | 0.28 | 0.42 | 0 | 0 |
| AvgConnBl_addition | 0.28 | 0.18 | 0.64 | 0.1 | 2.79 | 0.2 | 0 | 0 |
| WdPolysemyCnt | 0.28 | 0.33 | 0.11 | 0.6 | 0.09 | 0.39 | 0 | 0 |
| AvgAOABl_Bristol | 0.28 | 0.19 | 0.22 | 0.38 | 0.49 | 0.43 | 0 | 0 |
| AvgNmdEntSen | 0.27 | 0.26 | 0.54 | 0.39 | 0.36 | 0.19 | 0 | 0 |
| AvgAOEBl_InfPointPoly | 0.27 | 0.2 | 0.35 | 0.23 | 2.11 | 0.3 | 0 | 0.76 |
| AvgConnSen_temp_conns | 0.27 | 0.35 | 0.12 | 0 | 0.93 | 0.48 | 0 | 0 |
| WdAvgDpthHypernymTree | 0.27 | 0.43 | 0.4 | 0.2 | 0.15 | 0.25 | 0 | 0 |
| AvgDepsSen_dep | 0.27 | 0.06 | 0.38 | 0.28 | 2.35 | 0.34 | 0 | 0.3 |
| AvgAOESen_InvLinRegSlo | 0.26 | 0.27 | 0.52 | 0.25 | 0.02 | 0.19 | 0 | 0.3 |
| AvgAOASen_Bird | 0.26 | 0.53 | 0.04 | 0.16 | 0.75 | 0.39 | 0 | 0.15 |
| AvgNmdEntBl | 0.26 | 0.17 | 0.48 | 0.04 | 1.53 | 0.3 | 0 | 0 |
| AvgConnSen_reas_purp | 0.25 | 0.12 | 0.08 | 0.48 | 1.14 | 0.4 | 0 | 0 |
| AvgDepsSen_det | 0.25 | 0.11 | 0.1 | 0.25 | 0.26 | 0.44 | 0 | 0.76 |
| AvgAOABl_Bird | 0.23 | 0.7 | 0.21 | 0.33 | 0.44 | 0.13 | 0 | 0 |
| AvgAOESen_IndexPolyFAT.3 | 0.23 | 0.11 | 0.28 | 0.31 | 0.64 | 0.31 | 0 | 0 |
| AvgDepsBl_dep | 0.22 | 0.1 | 0.44 | 0.05 | 1.31 | 0.22 | 0 | 0.61 |
| AvgDepsBl_nsubjpass | 0.21 | 0.02 | 0.84 | 0.2 | 0.07 | 0 | 0 | 0 |
| AvgAOESen_IndexAbThr.0.3. | 0.19 | 0.27 | 0.12 | 0.39 | 0.05 | 0.22 | 0 | 0 |
| AvgDepsSen_aux | 0.18 | 0.39 | 0 | 0.49 | 2.05 | 0.17 | 0 | 0 |
| AvgUnqWdBl | 0.15 | 0 | 0 | 1.66 | 0 | 0 | 0 | 0 |
| AvgBlLen | 0.15 | 0 | 0 | 1.68 | 0 | 0 | 0 | 0 |
| AvgDepsBl_acl | 0.14 | 0.01 | 0.42 | 0.04 | 0.2 | 0.1 | 0 | 0 |
| AvgVerbBl | 0.14 | 0 | 0 | 1.57 | 0 | 0 | 0 | 0 |
| Content.words | 0.14 | 0 | 0 | 1.57 | 0 | 0 | 0 | 0 |
| AvgWdBl | 0.14 | 0 | 0 | 1.57 | 0 | 0 | 0 | 0 |
| AvgNounNmdEntBl | 0.14 | 0.29 | 0.05 | 0 | 1.08 | 0.21 | 0 | 0 |
| Words | 0.13 | 0 | 0 | 1.47 | 0 | 0 | 0 | 0 |
| AvgPrepositionBl | 0.11 | 0 | 0 | 1.22 | 0 | 0 | 0 | 0 |
| AvgDepsSen_advcl | 0.1 | 0.14 | 0.05 | 0.52 | 0.09 | 0.05 | 0 | 0 |
| AvgDepsBl_case | 0.09 | 0 | 0 | 0.96 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_LDA | 0.09 | 0 | 0 | 0.99 | 0 | 0 | 0 | 0 |
| LangRhythmId | 0.09 | 0.01 | 0.19 | 0 | 0.98 | 0.1 | 0 | 0 |
| AvgIntraBlCoh_Path | 0.08 | 0 | 0 | 0.84 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LDA | 0.08 | 0 | 0 | 0.87 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_LSA | 0.08 | 0 | 0 | 0.91 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_LSA | 0.07 | 0 | 0 | 0.74 | 0 | 0 | 0 | 0 |
| SenScoreStDev | 0.07 | 0 | 0 | 0.75 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_Path | 0.07 | 0 | 0 | 0.76 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_word2vec | 0.07 | 0 | 0 | 0.79 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_Path | 0.07 | 0 | 0 | 0.81 | 0 | 0 | 0 | 0 |
| AvgNounBl | 0.07 | 0 | 0 | 0.83 | 0 | 0 | 0 | 0 |
| Sentences | 0.07 | 0 | 0 | 0.83 | 0 | 0 | 0 | 0 |
| AvgSenBl | 0.07 | 0 | 0 | 0.83 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LSA | 0.07 | 0 | 0 | 0.84 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_WuPalmer | 0.06 | 0 | 0 | 0.64 | 0 | 0 | 0 | 0 |
| SenStDevUnqWd | 0.06 | 0 | 0 | 0.66 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_LeackChod | 0.06 | 0 | 0 | 0.66 | 0 | 0 | 0 | 0 |
| AvgSenAdjCoh_LeackChod | 0.06 | 0 | 0 | 0.66 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Shock | 0.06 | 0 | 0 | 0.67 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_WuPalmer | 0.06 | 0 | 0 | 0.68 | 0 | 0 | 0 | 0 |
| AvgSenBlCoh_WuPalmer | 0.06 | 0 | 0 | 0.69 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_LeackChod | 0.06 | 0 | 0 | 0.69 | 0 | 0 | 0 | 0 |
| AvgIntraBlCoh_word2vec | 0.06 | 0 | 0 | 0.7 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Cortese | 0.06 | 0 | 0 | 0.7 | 0 | 0 | 0 | 0 |
| AvgVoice | 0.05 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0 |
| AvgConnSen_sentence_link | 0.05 | 0 | 0 | 0.51 | 0 | 0 | 0 | 0 |
| AvgVerbSen | 0.05 | 0 | 0 | 0.51 | 0 | 0 | 0 | 0 |
| AvgDepsSen_nsubj | 0.05 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Kuperman | 0.05 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 |
| AvgConnSen_conjunctions | 0.05 | 0 | 0 | 0.54 | 0 | 0 | 0 | 0 |
| AvgConnSen_coord_connects | 0.05 | 0 | 0 | 0.54 | 0 | 0 | 0 | 0 |
| AvgAdjectiveBl | 0.05 | 0 | 0 | 0.56 | 0 | 0 | 0 | 0 |
| AvgDepsSen_case | 0.05 | 0 | 0 | 0.56 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_IndexPolyFAT.3 | 0.04 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 |
| AvgPrepositionSen | 0.04 | 0 | 0 | 0.41 | 0 | 0 | 0 | 0 |
| AvgAdverbSen | 0.04 | 0 | 0 | 0.43 | 0 | 0 | 0 | 0 |
| AvgSenSyll | 0.04 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_IndexAbThr.0.3. | 0.04 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgAOEBl_IndexAbThr.0.3. | 0.04 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgConnSen_addition | 0.04 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgSemDep | 0.04 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgDepsSen_cc | 0.04 | 0 | 0 | 0.45 | 0 | 0 | 0 | 0 |
| AvgDepsBl_advmod | 0.04 | 0 | 0 | 0.46 | 0 | 0 | 0 | 0 |
| AvgWdSen | 0.03 | 0 | 0 | 0.29 | 0 | 0 | 0 | 0 |
| AvgSenStressedSyll | 0.03 | 0 | 0 | 0.3 | 0 | 0 | 0 | 0 |
| AvgConnSen_contrasts | 0.03 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 |
| AvgSenScore | 0.03 | 0 | 0 | 0.31 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InvLinRegSlo | 0.03 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Bird | 0.03 | 0 | 0 | 0.33 | 0 | 0 | 0 | 0 |
| AvgConnSen_coord_conjs | 0.03 | 0 | 0 | 0.34 | 0 | 0 | 0 | 0 |
| AvgDepsSen_conj | 0.03 | 0 | 0 | 0.35 | 0 | 0 | 0 | 0 |
| SynSoph | 0.03 | 0 | 0 | 0.38 | 0 | 0 | 0 | 0 |
| AvgAOADoc_Bristol | 0.03 | 0 | 0 | 0.38 | 0 | 0 | 0 | 0 |
| AvgAdverbBl | 0.03 | 0 | 0 | 0.38 | 0 | 0 | 0 | 0 |
| AvgAOESen_InvAverage | 0.02 | 0 | 0 | 0.17 | 0 | 0 | 0 | 0 |
| AvgConnBl_coord_connects | 0.02 | 0 | 0 | 0.2 | 0 | 0 | 0 | 0 |
| AvgDepsBl_auxpass | 0.02 | 0 | 0 | 0.22 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InfPointPoly | 0.02 | 0 | 0 | 0.23 | 0 | 0 | 0 | 0 |
| WdMaxDpthHypernymTree | 0.02 | 0 | 0 | 0.23 | 0 | 0 | 0 | 0 |
| AvgAOEDoc_InvAverage | 0.02 | 0 | 0 | 0.23 | 0 | 0 | 0 | 0 |
| AvgAOEBl_InvAverage | 0.02 | 0 | 0 | 0.23 | 0 | 0 | 0 | 0 |
| RdbltyKincaid | 0.02 | 0 | 0 | 0.24 | 0 | 0 | 0 | 0 |
| AvgUnqWdSen | 0.02 | 0 | 0 | 0.25 | 0 | 0 | 0 | 0 |
| RdbltyFog | 0.02 | 0 | 0 | 0.27 | 0 | 0 | 0 | 0 |
| AvgRhythmUnitSyll | 0.02 | 0 | 0 | 0.27 | 0 | 0 | 0 | 0 |
| AvgSenLen | 0.02 | 0 | 0 | 0.28 | 0 | 0 | 0 | 0 |
| AvgDepsBl_conj | 0.01 | 0 | 0 | 0.11 | 0 | 0 | 0 | 0 |
| AvgConnBl_conjunctions | 0.01 | 0 | 0 | 0.13 | 0 | 0 | 0 | 0 |
| AvgDepsBl_cc | 0.01 | 0 | 0 | 0.14 | 0 | 0 | 0 | 0 |
| AvgConnBl_coord_conjs | 0.01 | 0 | 0 | 0.16 | 0 | 0 | 0 | 0 |
| AvgUnqNmdEntBl | 0 | 0 | 0 | 0.05 | 0 | 0 | 0 | 0 |
ReaderBench Model 1d
This model was trained on principal component scores for fall data in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -8.4195 | 0.0406 | 0.8127 | 0.0694 | -0.0509 | 0.1058 | 0.0038 | 0.0448 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
PC1 = scores on 1st principal component extracted, …
Note: Importance is unavailable for support vector machines when PCA-based pre-processing is used (so all values for svm are 0).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| PC2 | 50.2 | 61.92 | 52.85 | 0 | 12.57 | 36.4 | 46.85 | 23.81 |
| PC1 | 8.03 | 1.95 | 9.25 | 0 | 1.38 | 5.78 | 0.27 | 8.39 |
| PC3 | 7.38 | 1.56 | 8.43 | 0 | 3.57 | 5.47 | 0 | 8.39 |
| PC5 | 5.42 | 3.46 | 5.74 | 0 | 5.67 | 3.71 | 0 | 13.83 |
| PC4 | 2 | 1.81 | 2.27 | 0 | 1.61 | 1.06 | 0 | 0 |
| PC24 | 1.9 | 0.9 | 1.48 | 0 | 6.31 | 1.73 | 16.76 | 1.59 |
| PC14 | 1.7 | 1.4 | 1.56 | 0 | 3.22 | 1.85 | 3.63 | 3.85 |
| PC8 | 1.47 | 0.85 | 1.45 | 0 | 1.88 | 2.26 | 0 | 1.59 |
| PC30 | 1.47 | 1.67 | 1.1 | 0 | 6.24 | 2.51 | 0 | 6.58 |
| PC6 | 1.09 | 0.73 | 0.95 | 0 | 0.78 | 2.84 | 0 | 0 |
| PC31 | 1.09 | 1.01 | 0.92 | 0 | 5.49 | 0.03 | 0 | 9.98 |
| PC7 | 1.03 | 0.17 | 1.17 | 0 | 1.27 | 0.95 | 0 | 0 |
| PC17 | 1.02 | 1.53 | 0.62 | 0 | 1.24 | 3.12 | 0 | 4.31 |
| PC43 | 0.94 | 1.87 | 0.58 | 0 | 5.88 | 1.5 | 0 | 4.76 |
| PC33 | 0.87 | 0.6 | 0.88 | 0 | 5.75 | 0.44 | 0 | 0 |
| PC32 | 0.82 | 0.21 | 0.6 | 0 | 3.29 | 2.36 | 0 | 1.59 |
| PC39 | 0.81 | 1.14 | 0.54 | 0 | 4.22 | 1.87 | 1.81 | 0 |
| PC27 | 0.77 | 0.48 | 0.66 | 0 | 2.74 | 0.62 | 0 | 4.99 |
| PC13 | 0.73 | 1.28 | 0.73 | 0 | 1.02 | 0.55 | 0 | 0 |
| PC34 | 0.7 | 0.46 | 0.2 | 0 | 0.43 | 2.05 | 13.64 | 0 |
| PC38 | 0.67 | 0.36 | 0.55 | 0 | 4.08 | 0.86 | 0 | 2.72 |
| PC23 | 0.64 | 0.5 | 0.72 | 0 | 2.45 | 0.1 | 0 | 0 |
| PC19 | 0.6 | 0.9 | 0.56 | 0 | 1.28 | 0.84 | 0 | 0 |
| PC16 | 0.6 | 0.67 | 0.22 | 0 | 0 | 2.25 | 6.99 | 0 |
| PC45 | 0.6 | 1.92 | 0.07 | 0 | 0 | 1.77 | 10.06 | 0.91 |
| PC20 | 0.56 | 0 | 0.53 | 0 | 1.3 | 1.26 | 0 | 0 |
| PC28 | 0.54 | 1.55 | 0.39 | 0 | 1.29 | 0.94 | 0 | 0.45 |
| PC40 | 0.54 | 0.77 | 0.52 | 0 | 4.01 | 0.04 | 0 | 0.45 |
| PC18 | 0.53 | 0.36 | 0.47 | 0 | 0.92 | 1.27 | 0 | 0 |
| PC11 | 0.51 | 0.87 | 0.55 | 0 | 0.48 | 0.18 | 0 | 0 |
| PC22 | 0.49 | 0.35 | 0.42 | 0 | 0.99 | 1.2 | 0 | 0 |
| PC36 | 0.46 | 0.95 | 0.09 | 0 | 0 | 3.14 | 0 | 0 |
| PC12 | 0.45 | 0.22 | 0.46 | 0 | 0.33 | 0.68 | 0 | 0 |
| PC29 | 0.44 | 0.95 | 0.29 | 0 | 0.79 | 1.36 | 0 | 0 |
| PC41 | 0.43 | 0.27 | 0.35 | 0 | 2.57 | 0.79 | 0 | 0 |
| PC42 | 0.4 | 0.32 | 0.31 | 0 | 2.25 | 1.03 | 0 | 0 |
| PC9 | 0.37 | 0.34 | 0.4 | 0 | 0.13 | 0.33 | 0 | 0 |
| PC44 | 0.29 | 0.31 | 0.22 | 0 | 1.53 | 0.68 | 0 | 0 |
| PC46 | 0.28 | 0.91 | 0.05 | 0 | 0 | 1.65 | 0 | 0.45 |
| PC15 | 0.26 | 0.87 | 0.14 | 0 | 0 | 0.59 | 0 | 1.36 |
| PC26 | 0.25 | 0.16 | 0.29 | 0 | 0.59 | 0 | 0 | 0 |
| PC35 | 0.23 | 0.02 | 0.19 | 0 | 0.43 | 0.7 | 0 | 0 |
| PC37 | 0.13 | 0.09 | 0.13 | 0 | 0.03 | 0.22 | 0 | 0 |
| PC10 | 0.12 | 0.4 | 0 | 0 | 0 | 0.99 | 0 | 0 |
| PC25 | 0.08 | 0.29 | 0.08 | 0 | 0 | 0.03 | 0 | 0 |
| PC21 | 0.06 | 0.64 | 0.03 | 0 | 0 | 0.01 | 0 | 0 |
Proportion of Variance by Varimax Rotated Component (RC)
Due to space limitations, loadings for only the first five principal components are displayed.
| Variable | RC1 | RC2 | RC3 | RC5 | RC4 |
|---|---|---|---|---|---|
| SS loadings | 44.56 | 31.07 | 19.29 | 10.07 | 9.38 |
| Proportion Var | 0.22 | 0.15 | 0.10 | 0.05 | 0.05 |
| Cumulative Var | 0.22 | 0.38 | 0.47 | 0.52 | 0.57 |
| Proportion Explained | 0.39 | 0.27 | 0.17 | 0.09 | 0.08 |
| Cumulative Proportion | 0.39 | 0.66 | 0.83 | 0.92 | 1.00 |
Varimax Rotated Loadings
| Metric | RC1 | RC2 | RC3 | RC5 | RC4 |
|---|---|---|---|---|---|
| Sentences | -0.589 | 0.59 | -0.023 | -0.072 | -0.157 |
| Words | 0.086 | 0.947 | 0.097 | 0.203 | 0.039 |
| Content.words | -0.006 | 0.908 | 0.091 | 0.119 | 0.153 |
| RdbltyFlesch | -0.891 | -0.159 | -0.034 | -0.06 | 0.033 |
| RdbltyFog | 0.95 | 0.117 | 0.07 | 0.127 | -0.022 |
| RdbltyKincaid | 0.948 | 0.116 | 0.044 | 0.128 | -0.02 |
| RdbltyDaleChall | 0.4 | -0.276 | -0.098 | 0.136 | -0.503 |
| AvgBlLen | -0.041 | 0.903 | 0.095 | 0.055 | 0.156 |
| AvgCommaBl | -0.1 | 0.309 | 0.05 | -0.227 | 0.006 |
| AvgSenLen | 0.91 | 0.198 | 0.058 | 0.044 | 0.17 |
| AvgSenBl | -0.589 | 0.59 | -0.023 | -0.072 | -0.157 |
| AvgUnqWdBl | -0.015 | 0.901 | 0.103 | 0.081 | 0.118 |
| AvgUnqWdSen | 0.922 | 0.161 | 0.089 | 0.096 | 0.134 |
| AvgWdLen | -0.089 | 0.407 | 0.562 | -0.271 | 0.362 |
| AvgWdBl | -0.006 | 0.908 | 0.091 | 0.119 | 0.153 |
| AvgWdSen | 0.931 | 0.147 | 0.057 | 0.086 | 0.135 |
| CharEnt | -0.006 | 0.489 | 0.287 | -0.198 | 0.059 |
| SenStDevUnqWd | -0.429 | 0.328 | 0.099 | 0.075 | 0.453 |
| SenStdDevWd | -0.354 | 0.326 | 0.076 | 0.112 | 0.472 |
| WdEnt | 0.01 | 0.886 | 0.21 | 0.066 | 0.085 |
| WdLettStdDev | -0.158 | 0.401 | 0.362 | -0.257 | 0.224 |
| LxcDiv | 0.065 | 0.791 | 0.172 | 0.047 | 0.204 |
| LxcSoph | 0.334 | 0.207 | 0.6 | -0.068 | 0.407 |
| SynSoph | 0.825 | 0.348 | 0.092 | 0.118 | 0.259 |
| AvgNounBl | -0.017 | 0.705 | 0.149 | 0.361 | 0.031 |
| AvgPronounBl | 0.159 | 0.77 | 0.042 | 0.026 | -0.004 |
| AvgVerbBl | 0.102 | 0.911 | 0.04 | 0.051 | 0.058 |
| AvgAdverbBl | 0.043 | 0.697 | 0.012 | -0.126 | -0.103 |
| AvgAdjectiveBl | -0.011 | 0.517 | 0.115 | 0.028 | 0.031 |
| AvgPrepositionBl | 0.078 | 0.786 | 0.06 | 0.154 | -0.004 |
| AvgNounSen | 0.834 | -0.018 | 0.109 | 0.357 | -0.019 |
| AvgPronounSen | 0.928 | 0.03 | 0.053 | 0.017 | -0.023 |
| AvgVerbSen | 0.957 | 0.1 | 0.031 | 0.013 | 0.042 |
| AvgAdverbSen | 0.701 | 0.22 | -0.042 | -0.147 | -0.05 |
| AvgAdjectiveSen | 0.699 | 0.007 | 0.137 | 0.004 | -0.055 |
| AvgPrepositionSen | 0.803 | 0.212 | 0.018 | 0.139 | -0.013 |
| AvgUnqNoundBl | -0.014 | 0.632 | 0.13 | 0.431 | -0.023 |
| AvgUnqPronounBl | -0.007 | 0.533 | 0.088 | 0.074 | 0.058 |
| AvgUnqVerbBl | 0.122 | 0.858 | 0.047 | 0.038 | 0.043 |
| AvgUnqAdverbBl | -0.007 | 0.719 | 0.011 | -0.17 | -0.142 |
| AvgUnqAdjectiveBl | -0.025 | 0.523 | 0.104 | -0.022 | 0.022 |
| AvgUnqPrepositionBl | 0.054 | 0.813 | 0.061 | 0.075 | -0.009 |
| AvgPronBl_first_person | 0.168 | 0.637 | 0.017 | 0.058 | -0.022 |
| AvgPronBl_indefinite | 0.011 | 0.577 | 0.018 | 0.033 | 0.138 |
| AggPronSen_indefinite | 0.677 | 0.159 | -0.005 | 0.004 | 0.112 |
| AvgPronBl_third_person | 0.131 | 0.424 | 0.057 | -0.018 | 0.019 |
| AggPronSen_third_person | 0.782 | -0.078 | 0.043 | -0.033 | 0.007 |
| AvgSemDep | 0.967 | 0.07 | 0.088 | 0.181 | 0.001 |
| WdDiffLemmaStem | -0.016 | 0.289 | 0.104 | 0.007 | -0.106 |
| WdDiffWdStem | 0.041 | 0.483 | 0.031 | -0.313 | 0.132 |
| WdMaxDpthHypernymTree | -0.08 | 0.083 | 0.356 | 0.059 | 0.349 |
| WdAvgDpthHypernymTree | -0.064 | 0.073 | 0.369 | 0.063 | 0.349 |
| WdPathCntHypernymTree | -0.066 | 0.123 | 0.217 | -0.019 | 0.464 |
| WdPolysemyCnt | 0 | -0.045 | -0.037 | 0.192 | 0.244 |
| WdSylCnt | -0.126 | 0.215 | 0.592 | -0.117 | 0.108 |
| AvgAOADoc_Shock | -0.084 | 0.426 | 0.396 | 0.15 | 0.04 |
| AvgAOABl_Shock | -0.084 | 0.426 | 0.396 | 0.15 | 0.04 |
| AvgAOASen_Shock | 0.289 | 0.196 | 0.419 | 0.105 | 0.111 |
| AvgAOADoc_Cortese | 0.011 | 0.056 | 0.743 | -0.088 | 0.221 |
| AvgAOABl_Cortese | 0.011 | 0.056 | 0.743 | -0.088 | 0.221 |
| AvgAOASen_Cortese | 0.261 | 0.168 | 0.575 | 0.045 | 0.211 |
| AvgAOADoc_Kuperman | -0.016 | 0.083 | 0.785 | 0.109 | -0.07 |
| AvgAOABl_Kuperman | -0.016 | 0.083 | 0.785 | 0.109 | -0.07 |
| AvgAOASen_Kuperman | 0.1 | 0.18 | 0.742 | 0.068 | -0.023 |
| AvgAOADoc_Bird | 0.011 | 0.057 | 0.76 | -0.004 | 0.135 |
| AvgAOABl_Bird | 0.011 | 0.057 | 0.76 | -0.004 | 0.135 |
| AvgAOASen_Bird | 0.262 | 0.14 | 0.554 | 0.029 | 0.173 |
| AvgAOADoc_Bristol | 0.018 | 0.308 | 0.513 | 0.022 | 0.201 |
| AvgAOABl_Bristol | 0.018 | 0.308 | 0.513 | 0.022 | 0.201 |
| AvgAOASen_Bristol | 0.413 | 0.102 | 0.453 | 0.113 | 0.185 |
| AvgAOEDoc_IndexPolyFitAbThr.0.3. | -0.049 | 0.027 | 0.591 | 0.176 | -0.597 |
| AvgAOEBl_IndexPolyFitAbThr.0.3. | -0.049 | 0.027 | 0.591 | 0.176 | -0.597 |
| AvgAOESen_IndexPolyFitAbThr.0.3. | 0.006 | 0.133 | 0.617 | 0.152 | -0.495 |
| AvgAOEDoc_InverseLinearRegressionSlope | -0.082 | 0.015 | 0.857 | 0.089 | -0.285 |
| AvgAOEBl_InverseLinearRegressionSlope | -0.082 | 0.015 | 0.857 | 0.089 | -0.285 |
| AvgAOESen_InverseLinearRegressionSlope | 0.077 | 0.163 | 0.795 | 0.054 | -0.145 |
| AvgAOEDoc_InflectionPointPolynomial | -0.099 | 0.06 | 0.848 | -0.017 | -0.214 |
| AvgAOEBl_InflectionPointPolynomial | -0.099 | 0.06 | 0.848 | -0.017 | -0.214 |
| AvgAOESen_InflectionPointPolynomial | 0.064 | 0.198 | 0.8 | -0.021 | -0.076 |
| AvgAOEDoc_InverseAverage | -0.102 | 0.053 | 0.857 | -0.014 | -0.243 |
| AvgAOEBl_InverseAverage | -0.102 | 0.053 | 0.857 | -0.014 | -0.243 |
| AvgAOESen_InverseAverage | 0.06 | 0.191 | 0.808 | -0.021 | -0.097 |
| AvgAOEDoc_IndexAboveThreshold.0.3. | -0.093 | 0.045 | 0.476 | 0.179 | -0.636 |
| AvgAOEBl_IndexAboveThreshold.0.3. | -0.093 | 0.045 | 0.476 | 0.179 | -0.636 |
| AvgAOESen_IndexAboveThreshold.0.3. | -0.071 | 0.107 | 0.491 | 0.174 | -0.556 |
| AvgNmdEntBl | 0.057 | 0.52 | -0.052 | 0.139 | 0.032 |
| AvgNounNmdEntBl | 0.088 | 0.363 | -0.01 | 0.198 | -0.015 |
| AvgUnqNmdEntBl | 0.119 | 0.537 | -0.058 | 0.14 | -0.003 |
| AvgNmdEntSen | 0.752 | 0.106 | -0.084 | 0.167 | 0.06 |
| TCorefChainDoc | -0.03 | 0.621 | 0.189 | 0.003 | -0.062 |
| AvgCorefChain | 0.143 | 0.47 | 0.111 | 0.016 | 0.079 |
| AvgChainSpan | 0.088 | 0.729 | 0.038 | -0.003 | 0.128 |
| AvgInferenceDistChain | 0.245 | 0.306 | 0.046 | 0.001 | -0.012 |
| TActCorefChainWd | -0.092 | -0.329 | 0.268 | -0.225 | -0.102 |
| TCorefChainBigSpan | 0.108 | 0.426 | 0.243 | -0.098 | 0.002 |
| AvgConnBl_addition | 0.067 | 0.309 | 0.032 | 0.777 | 0.055 |
| AvgConnSen_addition | 0.658 | -0.166 | 0.072 | 0.622 | -0.077 |
| AvgConnBl_conjunctions | 0.168 | 0.362 | 0.061 | 0.775 | -0.017 |
| AvgConnSen_conjunctions | 0.72 | -0.147 | 0.097 | 0.579 | -0.152 |
| AvgConnBl_contrasts | 0.076 | 0.444 | 0.114 | 0.035 | -0.118 |
| AvgConnSen_contrasts | 0.512 | 0.196 | 0.125 | -0.059 | -0.149 |
| AvgConnBl_coordinating_conjuncts | 0.381 | 0.45 | -0.054 | 0.141 | 0.092 |
| AvgConnSen_coordinating_conjuncts | 0.72 | 0.205 | -0.012 | -0.003 | -0.013 |
| AvgConnBl_coordinating_connectives | 0.255 | 0.506 | 0.035 | 0.7 | 0.003 |
| AvgConnSen_coordinating_connectives | 0.818 | -0.034 | 0.071 | 0.48 | -0.119 |
| AvgConnBl_logical_connectors | 0.186 | 0.317 | 0.046 | 0.76 | -0.008 |
| AvgConnSen_logical_connectors | 0.693 | -0.154 | 0.069 | 0.563 | -0.121 |
| AvgConnBl_oppositions | 0.096 | 0.391 | 0.08 | 0.048 | -0.132 |
| AvgConnSen_oppositions | 0.494 | 0.14 | 0.106 | -0.083 | -0.187 |
| AvgConnBl_order | -0.115 | 0.255 | -0.013 | 0.188 | 0.176 |
| AvgConnSen_order | 0.315 | 0.084 | -0.013 | 0.115 | 0.215 |
| AvgConnBl_reason_and_purpose | 0.204 | 0.536 | -0.017 | 0.194 | 0.163 |
| AvgConnSen_reason_and_purpose | 0.71 | 0.226 | 0.002 | 0.034 | 0.064 |
| AvgConnBl_semi_coordinators | 0.381 | 0.45 | -0.054 | 0.141 | 0.092 |
| AvgConnSen_semi_coordinators | 0.72 | 0.205 | -0.012 | -0.003 | -0.013 |
| AvgConnBl_sentence_linking | 0.241 | 0.603 | 0.014 | 0.606 | 0.063 |
| AvgConnSen_sentence_linking | 0.862 | 0.027 | 0.048 | 0.411 | -0.059 |
| AvgConnBl_simple_subordinators | 0.081 | 0.52 | 0.062 | -0.082 | -0.029 |
| AvgConnSen_simple_subordinators | 0.681 | 0.165 | -0.053 | -0.039 | 0.023 |
| AvgConnBl_temporal_connectors | 0.121 | 0.382 | -0.081 | -0.07 | 0.114 |
| AvgConnSen_temporal_connectors | 0.637 | 0.153 | -0.183 | -0.036 | 0.106 |
| LexChainAvgSpan | 0.128 | 0.407 | 0.308 | 0.073 | 0.415 |
| LexChainMaxSp | -0.007 | 0.663 | 0.02 | 0.171 | 0.207 |
| AvgBlScore | 0.148 | 0.801 | 0.043 | 0.165 | 0.285 |
| AvgSenScore | 0.909 | 0.12 | 0.044 | 0.055 | 0.165 |
| SenScoreStDev | -0.466 | 0.332 | 0.071 | 0.09 | 0.501 |
| AvgIntraBlCoh_LeackockChodorow | -0.741 | 0.302 | 0.152 | 0.008 | 0.471 |
| AvgSenAdjCoh_LeackockChodorow | -0.735 | 0.263 | 0.166 | -0.002 | 0.488 |
| AvgSenBlCoh_LeackockChodorow | 0.434 | -0.139 | 0.696 | 0.053 | 0.24 |
| AvgIntraBlCoh_WuPalmer | -0.748 | 0.296 | 0.152 | 0.001 | 0.465 |
| AvgSenAdjCoh_WuPalmer | -0.743 | 0.259 | 0.167 | -0.01 | 0.482 |
| AvgSenBlCoh_WuPalmer | 0.443 | -0.145 | 0.694 | 0.049 | 0.226 |
| AvgIntraBlCoh_Path | -0.749 | 0.277 | 0.154 | -0.007 | 0.46 |
| AvgSenAdjCoh_Path | -0.744 | 0.234 | 0.175 | -0.021 | 0.478 |
| AvgSenBlCoh_Path | 0.528 | -0.219 | 0.642 | 0.044 | 0.155 |
| AvgIntraBlCoh_LSA | -0.739 | 0.338 | 0.148 | 0.005 | 0.446 |
| AvgSenAdjCoh_LSA | -0.729 | 0.295 | 0.166 | -0.011 | 0.473 |
| AvgSenBlCoh_LSA | 0.598 | -0.209 | 0.547 | 0.09 | 0.148 |
| AvgIntraBlCoh_LDA | -0.738 | 0.345 | 0.151 | -0.004 | 0.449 |
| AvgSenAdjCoh_LDA | -0.718 | 0.324 | 0.158 | -0.003 | 0.473 |
| AvgSenBlCoh_LDA | 0.513 | -0.066 | 0.598 | 0.08 | 0.202 |
| AvgIntraBlCoh_word2vec | -0.743 | 0.31 | 0.158 | -0.026 | 0.457 |
| AvgSenAdjCoh_word2vec | -0.734 | 0.273 | 0.179 | -0.046 | 0.48 |
| AvgSenBlCoh_word2vec | 0.581 | -0.259 | 0.577 | 0.048 | 0.165 |
| AvgBlVoiceCoOcc | 0.083 | 0.49 | -0.117 | 0.351 | 0.258 |
| AvgVoice | 0.086 | 0.506 | -0.117 | 0.355 | 0.247 |
| AvgSenSyll | 0.973 | 0.081 | 0.076 | 0.138 | -0.008 |
| AvgSenStressedSyll | 0.939 | 0.141 | 0.049 | 0.106 | 0.117 |
| AvgRhythmUnits | 0.315 | 0.184 | -0.003 | -0.262 | 0.043 |
| AvgRhythmUnitSyll | 0.807 | -0.027 | 0.082 | 0.284 | -0.045 |
| AvgRhythmUnitStreesSyll | 0.809 | 0.024 | 0.053 | 0.235 | 0.073 |
| LangRhythmCoeff | 0.24 | 0.048 | -0.011 | 0.129 | -0.121 |
| LangRhythmId | -0.173 | -0.023 | -0.06 | 0.028 | -0.387 |
| FrqRhythmId | 0.657 | -0.336 | -0.016 | 0.096 | 0.056 |
| LangRhythmDiameter | 0.373 | 0.113 | 0.117 | 0.197 | -0.139 |
| SenAsson | -0.051 | 0.221 | 0.079 | 0.066 | 0.065 |
| AvgDepsBl_acl | 0.092 | 0.23 | 0.074 | 0.206 | 0.093 |
| AvgDepsSen_acl | 0.485 | 0.081 | 0.031 | 0.343 | 0.108 |
| AvgDepsBl_advcl | 0.394 | 0.519 | -0.037 | 0.042 | 0.175 |
| AvgDepsSen_advcl | 0.768 | 0.219 | -0.041 | 0.009 | 0.065 |
| AvgDepsBl_advmod | 0.141 | 0.71 | -0.023 | -0.136 | -0.045 |
| AvgDepsSen_advmod | 0.753 | 0.193 | -0.054 | -0.132 | -0.006 |
| AvgDepsBl_amod | -0.092 | 0.476 | 0.104 | 0.038 | 0.13 |
| AvgDepsSen_amod | 0.563 | 0.072 | 0.117 | -0.001 | 0.112 |
| AvgDepsBl_aux | -0.02 | 0.402 | 0.09 | -0.282 | 0.005 |
| AvgDepsSen_aux | 0.621 | 0.072 | 0.112 | -0.18 | -0.071 |
| AvgDepsBl_auxpass | -0.079 | 0.424 | 0.001 | -0.052 | -0.166 |
| AvgDepsBl_case | -0.121 | 0.772 | 0.094 | 0.169 | -0.072 |
| AvgDepsSen_case | 0.747 | 0.175 | 0.093 | 0.178 | -0.024 |
| AvgDepsBl_cc | 0.203 | 0.37 | 0.063 | 0.746 | -0.039 |
| AvgDepsSen_cc | 0.73 | -0.143 | 0.088 | 0.553 | -0.149 |
| AvgDepsBl_ccomp | 0.383 | 0.496 | 0.036 | -0.022 | 0.173 |
| AvgDepsSen_ccomp | 0.781 | 0.073 | 0.031 | -0.041 | 0.09 |
| AvgDepsBl_compound | 0.104 | 0.149 | 0.038 | 0.366 | -0.006 |
| AvgDepsSen_compound | 0.587 | -0.062 | 0.073 | 0.412 | -0.064 |
| AvgDepsBl_conj | 0.31 | 0.297 | 0.122 | 0.722 | -0.045 |
| AvgDepsSen_conj | 0.731 | -0.134 | 0.134 | 0.536 | -0.162 |
| AvgDepsBl_cop | -0.001 | 0.419 | 0.055 | -0.12 | -0.002 |
| AvgDepsSen_cop | 0.583 | 0.061 | 0.018 | -0.107 | 0.003 |
| AvgDepsBl_dep | 0.234 | 0.123 | 0.104 | 0.47 | -0.059 |
| AvgDepsSen_dep | 0.527 | -0.187 | 0.15 | 0.494 | -0.174 |
| AvgDepsBl_det | -0.182 | 0.549 | 0.13 | 0.222 | 0.071 |
| AvgDepsSen_det | 0.576 | -0.06 | 0.152 | 0.415 | -0.023 |
| AvgDepsBl_dobj | 0.119 | 0.64 | 0.078 | 0.196 | 0.13 |
| AvgDepsSen_dobj | 0.856 | -0.002 | 0.047 | 0.128 | 0.028 |
| AvgDepsBl_mark | 0.367 | 0.535 | 0.01 | 0.087 | 0.139 |
| AvgDepsSen_mark | 0.795 | 0.152 | -0.063 | 0.085 | 0.073 |
| AvgDepsBl_mwe | -0.01 | 0.262 | 0.021 | 0.142 | 0.008 |
| AvgDepsSen_mwe | 0.383 | 0.097 | 0.025 | 0.044 | 0.031 |
| AvgDepsBl_neg | 0.197 | 0.361 | 0.053 | -0.181 | -0.132 |
| AvgDepsSen_neg | 0.549 | 0.073 | -0.024 | -0.17 | -0.086 |
| AvgDepsBl_nmod | -0.134 | 0.727 | 0.094 | 0.237 | -0.052 |
| AvgDepsSen_nmod | 0.661 | 0.15 | 0.086 | 0.281 | -0.02 |
| AvgDepsBl_nsubj | 0.141 | 0.88 | 0.062 | 0.041 | 0.08 |
| AvgDepsSen_nsubj | 0.969 | 0.02 | 0.051 | 0.018 | 0.003 |
| AvgDepsBl_nsubjpass | -0.015 | 0.417 | -0.005 | -0.065 | -0.158 |
| AvgDepsBl_nummod | 0.132 | 0.492 | -0.071 | -0.009 | 0.13 |
| AvgDepsBl_punct | -0.483 | 0.644 | -0.017 | -0.108 | -0.086 |
| AvgDepsSen_punct | -0.192 | 0.389 | 0.063 | -0.232 | 0.191 |
| AvgDepsBl_xcomp | 0.053 | 0.501 | 0.03 | 0.186 | 0.068 |
| AvgDepsSen_xcomp | 0.666 | 0.075 | 0.061 | 0.196 | 0.024 |
ReaderBench Model 1e
This model was trained on principal component scores for winter data in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -8.0185 | 0.0573 | 0.5839 | 0.5269 | -0.3984 | 0.1184 | 0.1066 | 0.0459 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
PC1 = scores on 1st principal component extracted, …
Note: Importance is unavailable for support vector machines when PCA-based pre-processing is used (so all values for svm are 0).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| PC2 | 32.8 | 53 | 47.34 | 0 | 9.95 | 26.16 | 39.14 | 22.22 |
| PC1 | 7.63 | 1.98 | 13.73 | 0 | 1.84 | 4.83 | 0 | 11.11 |
| PC39 | 4.81 | 1.96 | 1.08 | 0 | 7.68 | 2.76 | 17.37 | 8.89 |
| PC5 | 4.35 | 2.14 | 4.78 | 0 | 4.24 | 3.9 | 0 | 13.33 |
| PC11 | 4.33 | 1.43 | 3.55 | 0 | 5.85 | 1.52 | 4.87 | 11.11 |
| PC37 | 4.21 | 1.94 | 1.15 | 0 | 7.47 | 1.75 | 12.15 | 6.67 |
| PC6 | 3.45 | 3.05 | 3.87 | 0 | 3.53 | 2.17 | 0 | 8.89 |
| PC26 | 3.28 | 2.41 | 1.25 | 0 | 4.41 | 1.69 | 14.49 | 0 |
| PC38 | 3.13 | 0.57 | 1.08 | 0 | 7.55 | 0.99 | 0 | 6.67 |
| PC14 | 2.89 | 4.55 | 1.94 | 0 | 3.33 | 6.23 | 0 | 6.67 |
| PC24 | 2.2 | 1.36 | 1.07 | 0 | 3.24 | 0.49 | 8.2 | 0 |
| PC40 | 2.12 | 0.85 | 0.72 | 0 | 5.11 | 1.28 | 0 | 2.22 |
| PC9 | 1.75 | 0.72 | 2.06 | 0 | 2.21 | 0.62 | 0 | 2.22 |
| PC33 | 1.63 | 0.91 | 0.67 | 0 | 2.86 | 2.41 | 2.5 | 0 |
| PC45 | 1.61 | 0.26 | 0.5 | 0 | 4.33 | 0.63 | 0 | 0 |
| PC44 | 1.51 | 0.65 | 0.49 | 0 | 3.6 | 1.94 | 0 | 0 |
| PC32 | 1.3 | 0.87 | 0.67 | 0 | 2.61 | 1.92 | 0 | 0 |
| PC20 | 1.29 | 1.94 | 1 | 0 | 2.28 | 0.76 | 0 | 0 |
| PC4 | 1.16 | 1.23 | 1.61 | 0 | 0.82 | 1.5 | 0 | 0 |
| PC19 | 1.07 | 1.79 | 0.78 | 0 | 1.41 | 2.19 | 0 | 0 |
| PC34 | 0.98 | 1.11 | 0.5 | 0 | 1.9 | 1.2 | 0.26 | 0 |
| PC43 | 0.97 | 0.27 | 0.42 | 0 | 2.53 | 0 | 0 | 0 |
| PC8 | 0.95 | 0.61 | 1.17 | 0 | 0.8 | 1.73 | 0 | 0 |
| PC15 | 0.94 | 1.05 | 0.88 | 0 | 1.21 | 1.47 | 0 | 0 |
| PC23 | 0.9 | 0.73 | 0.64 | 0 | 1.33 | 1.89 | 0 | 0 |
| PC17 | 0.9 | 0.77 | 0.85 | 0 | 1.41 | 0.6 | 0 | 0 |
| PC35 | 0.86 | 1.17 | 0.46 | 0 | 1.61 | 1.24 | 0 | 0 |
| PC3 | 0.81 | 0.64 | 1.28 | 0 | 0.16 | 1.75 | 0 | 0 |
| PC12 | 0.76 | 0.42 | 0.78 | 0 | 0.71 | 1.86 | 0 | 0 |
| PC28 | 0.73 | 0.92 | 0.48 | 0 | 1.01 | 1.9 | 0 | 0 |
| PC16 | 0.69 | 0.41 | 0.71 | 0 | 0.86 | 0.92 | 0 | 0 |
| PC25 | 0.59 | 0.21 | 0.47 | 0 | 0.73 | 1.59 | 0 | 0 |
| PC30 | 0.42 | 1.15 | 0.31 | 0 | 0.25 | 1.74 | 0 | 0 |
| PC42 | 0.41 | 0.61 | 0.22 | 0 | 0.34 | 1.92 | 0 | 0 |
| PC36 | 0.36 | 0.14 | 0.29 | 0 | 0.51 | 0.88 | 0 | 0 |
| PC27 | 0.35 | 0.38 | 0.35 | 0 | 0.34 | 0.77 | 0 | 0 |
| PC7 | 0.34 | 1.67 | 0.06 | 0 | 0 | 1.66 | 1.01 | 0 |
| PC13 | 0.28 | 0.89 | 0.01 | 0 | 0 | 2.56 | 0 | 0 |
| PC29 | 0.28 | 0.97 | 0.05 | 0 | 0 | 2.36 | 0 | 0 |
| PC10 | 0.27 | 0.54 | 0.27 | 0 | 0 | 1.27 | 0 | 0 |
| PC18 | 0.24 | 0.52 | 0.14 | 0 | 0 | 1.68 | 0 | 0 |
| PC31 | 0.18 | 0.35 | 0.08 | 0 | 0 | 1.51 | 0 | 0 |
| PC46 | 0.11 | 0.43 | 0.15 | 0 | 0 | 0.24 | 0 | 0 |
| PC21 | 0.07 | 0.18 | 0 | 0 | 0 | 0.64 | 0 | 0 |
| PC22 | 0.07 | 0.25 | 0.09 | 0 | 0 | 0.27 | 0 | 0 |
| PC41 | 0.06 | 0 | 0 | 0 | 0 | 0.6 | 0 | 0 |
Proportion of Variance by Varimax Rotated Component (RC)
Due to space limitations, loadings for only the first five principal components are displayed.
| Variable | RC1 | RC2 | RC3 | RC4 | RC5 |
|---|---|---|---|---|---|
| SS loadings | 46.99 | 34.24 | 14.94 | 7.95 | 6.95 |
| Proportion Var | 0.23 | 0.17 | 0.07 | 0.04 | 0.03 |
| Cumulative Var | 0.23 | 0.40 | 0.48 | 0.52 | 0.55 |
| Proportion Explained | 0.42 | 0.31 | 0.13 | 0.07 | 0.06 |
| Cumulative Proportion | 0.42 | 0.73 | 0.87 | 0.94 | 1.00 |
Varimax Rotated Loadings
| Metric | RC1 | RC2 | RC3 | RC4 | RC5 |
|---|---|---|---|---|---|
| Sentences | -0.66 | 0.48 | -0.05 | -0.04 | -0.26 |
| Words | 0.07 | 0.97 | 0 | -0.08 | -0.05 |
| Content.words | -0.05 | 0.94 | 0.07 | 0.07 | -0.11 |
| RdbltyFlesch | -0.82 | -0.19 | -0.02 | -0.12 | 0.09 |
| RdbltyFog | 0.91 | 0.17 | 0.01 | 0.04 | 0.01 |
| RdbltyKincaid | 0.91 | 0.18 | 0.01 | 0.05 | 0 |
| RdbltyDaleChall | 0.5 | -0.34 | 0.11 | -0.06 | -0.22 |
| AvgBlLen | -0.11 | 0.91 | 0.08 | 0.13 | -0.16 |
| AvgCommaBl | -0.23 | 0.38 | 0.06 | -0.1 | -0.1 |
| AvgSenLen | 0.9 | 0.26 | 0.08 | 0.15 | 0.03 |
| AvgSenBl | -0.66 | 0.48 | -0.05 | -0.04 | -0.26 |
| AvgUnqWdBl | -0.08 | 0.91 | 0.05 | 0.12 | -0.15 |
| AvgUnqWdSen | 0.93 | 0.22 | 0.08 | 0.12 | 0.05 |
| AvgWdLen | -0.21 | 0.26 | -0.17 | 0.25 | -0.46 |
| AvgWdBl | -0.05 | 0.94 | 0.07 | 0.07 | -0.11 |
| AvgWdSen | 0.93 | 0.23 | 0.07 | 0.09 | 0.06 |
| CharEnt | -0.14 | 0.54 | 0.11 | -0.02 | -0.03 |
| SenStDevUnqWd | -0.23 | 0.5 | 0.04 | 0.09 | 0.41 |
| SenStdDevWd | -0.16 | 0.48 | 0.04 | 0.07 | 0.45 |
| WdEnt | 0.03 | 0.86 | 0.04 | 0.1 | -0.13 |
| WdLettStdDev | -0.09 | 0.4 | 0.37 | 0.24 | 0.04 |
| LxcDiv | 0 | 0.81 | 0.09 | 0.2 | -0.07 |
| LxcSoph | 0.36 | 0.09 | -0.28 | 0.11 | -0.35 |
| SynSoph | 0.82 | 0.44 | 0.12 | 0.14 | 0.12 |
| AvgNounBl | 0.01 | 0.76 | 0.11 | 0.03 | -0.35 |
| AvgPronounBl | 0.1 | 0.79 | -0.1 | -0.13 | 0.18 |
| AvgVerbBl | 0.03 | 0.91 | -0.05 | -0.02 | 0.05 |
| AvgAdverbBl | 0.09 | 0.63 | -0.01 | -0.22 | 0.11 |
| AvgAdjectiveBl | -0.06 | 0.55 | 0.12 | -0.07 | -0.09 |
| AvgPrepositionBl | 0.11 | 0.75 | -0.04 | 0.27 | -0.1 |
| AvgNounSen | 0.9 | 0.06 | 0.06 | 0.06 | -0.14 |
| AvgPronounSen | 0.91 | 0.1 | -0.08 | -0.02 | 0.15 |
| AvgVerbSen | 0.94 | 0.16 | -0.01 | 0.04 | 0.09 |
| AvgAdverbSen | 0.77 | 0.15 | -0.02 | -0.13 | 0.18 |
| AvgAdjectiveSen | 0.71 | 0.11 | 0.13 | -0.13 | 0.1 |
| AvgPrepositionSen | 0.85 | 0.2 | -0.07 | 0.26 | -0.04 |
| AvgUnqNoundBl | 0.02 | 0.71 | 0.05 | 0.02 | -0.36 |
| AvgUnqPronounBl | 0.09 | 0.66 | -0.01 | -0.09 | 0.02 |
| AvgUnqVerbBl | 0.03 | 0.85 | -0.05 | 0.01 | 0.04 |
| AvgUnqAdverbBl | -0.01 | 0.61 | -0.02 | -0.13 | 0.05 |
| AvgUnqAdjectiveBl | -0.06 | 0.57 | 0.12 | -0.09 | -0.13 |
| AvgUnqPrepositionBl | 0.09 | 0.71 | 0 | 0.3 | -0.14 |
| AvgPronBl_first_person | 0.09 | 0.69 | -0.05 | -0.14 | 0.12 |
| AvgPronBl_indefinite | -0.05 | 0.45 | 0.01 | 0.24 | -0.04 |
| AggPronSen_indefinite | 0.62 | 0.17 | 0.05 | 0.23 | 0.02 |
| AvgPronBl_third_person | 0.13 | 0.48 | -0.08 | -0.12 | 0.14 |
| AggPronSen_third_person | 0.84 | -0.02 | -0.09 | -0.09 | 0.12 |
| AvgSemDep | 0.97 | 0.12 | -0.01 | -0.06 | 0.06 |
| WdDiffLemmaStem | -0.11 | 0.06 | -0.23 | -0.01 | -0.24 |
| WdDiffWdStem | -0.28 | 0.19 | 0.03 | 0.16 | -0.28 |
| WdMaxDpthHypernymTree | -0.02 | -0.26 | -0.09 | 0.14 | -0.23 |
| WdAvgDpthHypernymTree | 0 | -0.27 | -0.11 | 0.13 | -0.24 |
| WdPathCntHypernymTree | -0.1 | -0.26 | -0.17 | 0.16 | -0.09 |
| WdPolysemyCnt | 0.06 | 0.11 | -0.09 | -0.09 | 0.35 |
| WdSylCnt | -0.1 | 0.05 | -0.26 | 0.11 | -0.46 |
| AvgAOADoc_Shock | 0.03 | 0.43 | 0.12 | 0.27 | -0.28 |
| AvgAOABl_Shock | 0.03 | 0.43 | 0.12 | 0.27 | -0.28 |
| AvgAOASen_Shock | 0.46 | 0.21 | 0.05 | 0.31 | -0.27 |
| AvgAOADoc_Cortese | -0.17 | 0.07 | 0.69 | 0.22 | 0.19 |
| AvgAOABl_Cortese | -0.17 | 0.07 | 0.69 | 0.22 | 0.19 |
| AvgAOASen_Cortese | 0.14 | 0.01 | 0.55 | 0.3 | 0.17 |
| AvgAOADoc_Kuperman | -0.22 | -0.03 | 0.43 | 0.31 | -0.38 |
| AvgAOABl_Kuperman | -0.22 | -0.03 | 0.43 | 0.31 | -0.38 |
| AvgAOASen_Kuperman | -0.03 | -0.04 | 0.42 | 0.41 | -0.3 |
| AvgAOADoc_Bird | -0.12 | 0.17 | 0.55 | 0.32 | 0.21 |
| AvgAOABl_Bird | -0.12 | 0.17 | 0.55 | 0.32 | 0.21 |
| AvgAOASen_Bird | 0.23 | 0.12 | 0.43 | 0.36 | 0.21 |
| AvgAOADoc_Bristol | -0.06 | 0.24 | 0.54 | 0.25 | -0.04 |
| AvgAOABl_Bristol | -0.06 | 0.24 | 0.54 | 0.25 | -0.04 |
| AvgAOASen_Bristol | 0.37 | 0.08 | 0.34 | 0.26 | 0 |
| AvgAOEDoc_IndexPolyFAT.3 | -0.02 | -0.07 | 0.77 | -0.22 | -0.14 |
| AvgAOEBl_IndexPolyFAT.3 | -0.02 | -0.07 | 0.77 | -0.22 | -0.14 |
| AvgAOESen_IndexPolyFAT.3 | 0.02 | -0.02 | 0.74 | -0.12 | -0.17 |
| AvgAOEDoc_InvLinRegSlo | 0 | -0.1 | 0.82 | -0.22 | -0.09 |
| AvgAOEBl_InvLinRegSlo | 0 | -0.1 | 0.82 | -0.22 | -0.09 |
| AvgAOESen_InvLinRegSlo | 0.17 | -0.02 | 0.67 | -0.02 | -0.11 |
| AvgAOEDoc_InfPointPoly | -0.1 | 0.01 | 0.86 | -0.15 | 0.1 |
| AvgAOEBl_InfPointPoly | -0.1 | 0.01 | 0.86 | -0.15 | 0.1 |
| AvgAOESen_InfPointPoly | 0.07 | 0.03 | 0.73 | 0.02 | 0.05 |
| AvgAOEDoc_InvAverage | -0.11 | 0 | 0.88 | -0.14 | 0.06 |
| AvgAOEBl_InvAverage | -0.11 | 0 | 0.88 | -0.14 | 0.06 |
| AvgAOESen_InvAverage | 0.06 | 0.02 | 0.74 | 0.03 | 0.02 |
| AvgAOEDoc_IndexAbThr.0.3. | 0.03 | -0.04 | 0.77 | -0.12 | -0.21 |
| AvgAOEBl_IndexAbThr.0.3. | 0.03 | -0.04 | 0.77 | -0.12 | -0.21 |
| AvgAOESen_IndexAbThr.0.3. | 0.05 | 0 | 0.75 | -0.04 | -0.27 |
| AvgNmdEntBl | -0.07 | 0.32 | 0.15 | -0.11 | -0.44 |
| AvgNounNmdEntBl | -0.02 | 0.24 | 0.24 | -0.13 | -0.43 |
| AvgUnqNmdEntBl | -0.07 | 0.34 | 0.12 | -0.09 | -0.48 |
| AvgNmdEntSen | 0.59 | 0.01 | 0.19 | -0.09 | -0.26 |
| TCorefChainDoc | 0.04 | 0.69 | 0.06 | -0.2 | -0.01 |
| AvgCorefChain | 0.02 | 0.46 | -0.18 | 0 | 0.11 |
| AvgChainSpan | 0.06 | 0.73 | -0.03 | 0.07 | -0.1 |
| AvgInferenceDistChain | 0.45 | 0.31 | -0.02 | 0.31 | 0.07 |
| TActCorefChainWd | -0.02 | -0.25 | -0.04 | -0.19 | 0.04 |
| TCorefChainBigSpan | 0.19 | 0.52 | -0.02 | -0.09 | -0.01 |
| AvgConnBl_addition | 0.1 | 0.53 | 0.1 | -0.58 | -0.08 |
| AvgConnSen_addition | 0.74 | 0.04 | 0.05 | -0.4 | -0.02 |
| AvgConnBl_conjunctions | 0.15 | 0.59 | 0.09 | -0.57 | 0.06 |
| AvgConnSen_conjunctions | 0.83 | 0.04 | 0.06 | -0.36 | 0.06 |
| AvgConnBl_contrasts | 0.19 | 0.37 | 0 | -0.11 | 0.28 |
| AvgConnSen_contrasts | 0.63 | 0.11 | 0.04 | -0.11 | 0.22 |
| AvgConnBl_coord_conjs | 0.17 | 0.47 | -0.17 | 0.14 | 0.23 |
| AvgConnSen_coord_conjs | 0.63 | 0.25 | -0.14 | 0.21 | 0.25 |
| AvgConnBl_coord_connects | 0.21 | 0.7 | -0.02 | -0.4 | 0.18 |
| AvgConnSen_coord_connects | 0.88 | 0.1 | -0.02 | -0.23 | 0.17 |
| AvgConnBl_logical_conn | 0.09 | 0.5 | 0.07 | -0.64 | 0.05 |
| AvgConnSen_logical_conn | 0.67 | -0.02 | 0.03 | -0.5 | 0.1 |
| AvgConnBl_oppositions | 0.19 | 0.37 | 0.05 | 0.02 | 0.22 |
| AvgConnSen_oppositions | 0.6 | 0.16 | 0.07 | 0.12 | 0.11 |
| AvgConnBl_order | 0.12 | 0.38 | 0.05 | -0.29 | -0.16 |
| AvgConnSen_order | 0.57 | 0.14 | 0.02 | -0.07 | -0.1 |
| AvgConnBl_reas_purp | 0.18 | 0.58 | -0.07 | -0.03 | 0.09 |
| AvgConnSen_reas_purp | 0.73 | 0.25 | -0.06 | 0.08 | 0.09 |
| AvgConnBl_semi_coords | 0.17 | 0.47 | -0.17 | 0.14 | 0.23 |
| AvgConnSen_semi_coords | 0.63 | 0.25 | -0.14 | 0.21 | 0.25 |
| AvgConnBl_sentence_link | 0.2 | 0.77 | -0.01 | -0.33 | 0.13 |
| AvgConnSen_sentence_link | 0.92 | 0.14 | -0.03 | -0.13 | 0.12 |
| AvgConnBl_simp_subords | -0.02 | 0.41 | -0.05 | 0.27 | 0.11 |
| AvgConnSen_simp_subords | 0.52 | 0.18 | -0.09 | 0.34 | 0.16 |
| AvgConnBl_temp_conn | -0.14 | 0.36 | 0.05 | -0.23 | 0.08 |
| AvgConnSen_temp_conn | 0.36 | 0.06 | 0.04 | -0.25 | 0.27 |
| LexChainAvgSpan | 0.07 | 0.49 | 0.03 | -0.01 | 0.16 |
| LexChainMaxSp | 0 | 0.72 | 0 | -0.02 | 0.08 |
| AvgBlScore | 0.14 | 0.82 | 0.01 | 0 | 0.13 |
| AvgSenScore | 0.9 | 0.22 | 0.02 | 0.04 | 0.14 |
| SenScoreStDev | -0.27 | 0.5 | 0.04 | 0.05 | 0.45 |
| AvgIntraBlCoh_LeackChod | -0.73 | 0.44 | 0.16 | 0.2 | 0.17 |
| AvgSenAdjCoh_LeackChod | -0.7 | 0.43 | 0.19 | 0.22 | 0.17 |
| AvgSenBlCoh_LeackChod | 0.77 | -0.38 | -0.13 | -0.08 | 0.09 |
| AvgIntraBlCoh_WuPalmer | -0.74 | 0.44 | 0.16 | 0.2 | 0.18 |
| AvgSenAdjCoh_WuPalmer | -0.7 | 0.43 | 0.19 | 0.22 | 0.17 |
| AvgSenBlCoh_WuPalmer | 0.79 | -0.39 | -0.13 | -0.09 | 0.08 |
| AvgIntraBlCoh_Path | -0.73 | 0.43 | 0.16 | 0.2 | 0.19 |
| AvgSenAdjCoh_Path | -0.7 | 0.42 | 0.19 | 0.22 | 0.2 |
| AvgSenBlCoh_Path | 0.82 | -0.42 | -0.12 | -0.13 | 0.08 |
| AvgIntraBlCoh_LSA | -0.73 | 0.46 | 0.16 | 0.18 | 0.18 |
| AvgSenAdjCoh_LSA | -0.7 | 0.44 | 0.2 | 0.22 | 0.18 |
| AvgSenBlCoh_LSA | 0.8 | -0.35 | -0.11 | -0.12 | 0.11 |
| AvgIntraBlCoh_LDA | -0.73 | 0.48 | 0.16 | 0.18 | 0.15 |
| AvgSenAdjCoh_LDA | -0.7 | 0.47 | 0.2 | 0.2 | 0.15 |
| AvgSenBlCoh_LDA | 0.75 | -0.21 | -0.13 | -0.11 | 0.08 |
| AvgIntraBlCoh_word2vec | -0.73 | 0.45 | 0.16 | 0.19 | 0.18 |
| AvgSenAdjCoh_word2vec | -0.7 | 0.44 | 0.2 | 0.22 | 0.18 |
| AvgSenBlCoh_word2vec | 0.8 | -0.39 | -0.1 | -0.11 | 0.11 |
| AvgBlVoiceCoOcc | 0.03 | 0.63 | -0.05 | -0.04 | -0.01 |
| AvgVoice | -0.01 | 0.6 | -0.08 | -0.05 | 0 |
| AvgSenSyll | 0.98 | 0.12 | 0.01 | 0.02 | 0.02 |
| AvgSenStressedSyll | 0.94 | 0.22 | 0.07 | 0.08 | 0.05 |
| AvgRhythmUnits | 0.23 | 0.26 | 0.15 | 0.01 | 0.28 |
| AvgRhythmUnitSyll | 0.84 | 0.01 | -0.09 | 0.02 | -0.09 |
| AvgRhythmUnitStreesSyll | 0.81 | 0.11 | -0.01 | 0.08 | -0.05 |
| LangRhythmCoeff | 0.39 | -0.1 | -0.21 | -0.17 | -0.06 |
| LangRhythmId | -0.11 | -0.02 | -0.4 | -0.27 | -0.14 |
| FrqRhythmId | 0.72 | -0.3 | 0.01 | -0.12 | 0.04 |
| LangRhythmDiameter | 0.27 | -0.01 | -0.34 | -0.25 | -0.19 |
| SenAsson | 0.13 | 0.25 | 0.12 | -0.11 | -0.17 |
| AvgDepsBl_acl | -0.03 | 0.24 | 0.15 | 0.19 | -0.16 |
| AvgDepsSen_acl | 0.44 | 0.03 | 0.26 | 0.09 | -0.13 |
| AvgDepsBl_advcl | 0.23 | 0.52 | -0.05 | 0.1 | 0.16 |
| AvgDepsSen_advcl | 0.67 | 0.21 | -0.08 | 0.2 | 0.09 |
| AvgDepsBl_advmod | 0.07 | 0.67 | 0.03 | -0.25 | 0.13 |
| AvgDepsSen_advmod | 0.78 | 0.15 | 0.02 | -0.17 | 0.2 |
| AvgDepsBl_amod | -0.07 | 0.39 | 0.16 | 0.06 | -0.2 |
| AvgDepsSen_amod | 0.62 | 0.1 | 0.17 | 0 | -0.01 |
| AvgDepsBl_aux | 0.13 | 0.37 | -0.13 | 0.13 | 0.24 |
| AvgDepsSen_aux | 0.56 | 0.02 | -0.14 | -0.01 | 0.27 |
| AvgDepsBl_auxpass | 0.06 | 0.26 | 0.08 | 0.05 | 0.03 |
| AvgDepsBl_case | 0.06 | 0.69 | 0.01 | 0.15 | -0.26 |
| AvgDepsSen_case | 0.84 | 0.13 | -0.02 | 0.15 | -0.17 |
| AvgDepsBl_cc | 0.17 | 0.59 | 0.08 | -0.61 | 0.09 |
| AvgDepsSen_cc | 0.8 | 0.02 | 0.05 | -0.42 | 0.11 |
| AvgDepsBl_ccomp | 0.37 | 0.51 | -0.03 | -0.01 | 0.32 |
| AvgDepsSen_ccomp | 0.78 | 0.16 | -0.01 | -0.01 | 0.19 |
| AvgDepsBl_compound | 0.17 | 0.16 | 0.19 | -0.05 | -0.36 |
| AvgDepsSen_compound | 0.61 | -0.12 | 0.18 | -0.07 | -0.27 |
| AvgDepsBl_conj | 0.23 | 0.52 | 0.06 | -0.58 | 0.14 |
| AvgDepsSen_conj | 0.77 | 0.02 | 0.04 | -0.4 | 0.13 |
| AvgDepsBl_cop | 0.04 | 0.49 | 0.09 | 0.03 | -0.1 |
| AvgDepsSen_cop | 0.71 | 0.16 | 0.14 | 0.04 | -0.03 |
| AvgDepsBl_dep | 0.2 | 0.22 | 0.08 | -0.34 | 0.07 |
| AvgDepsSen_dep | 0.56 | -0.05 | 0.1 | -0.31 | 0.11 |
| AvgDepsBl_det | -0.09 | 0.58 | -0.03 | 0.05 | -0.28 |
| AvgDepsSen_det | 0.86 | 0.1 | -0.02 | 0.04 | -0.16 |
| AvgDepsBl_dobj | 0 | 0.73 | 0.03 | -0.2 | 0.01 |
| AvgDepsSen_dobj | 0.91 | 0.07 | -0.01 | -0.04 | 0.1 |
| AvgDepsBl_mark | 0.22 | 0.63 | -0.11 | 0.28 | 0.09 |
| AvgDepsSen_mark | 0.78 | 0.25 | -0.1 | 0.28 | 0.05 |
| AvgDepsBl_mwe | -0.07 | 0.19 | 0.06 | 0 | -0.02 |
| AvgDepsSen_mwe | 0.3 | 0 | 0.03 | -0.09 | 0.05 |
| AvgDepsBl_neg | 0.03 | 0.37 | -0.08 | -0.03 | 0.25 |
| AvgDepsSen_neg | 0.38 | 0.12 | -0.04 | 0.05 | 0.28 |
| AvgDepsBl_nmod | 0.05 | 0.64 | -0.01 | 0.17 | -0.27 |
| AvgDepsSen_nmod | 0.83 | 0.07 | -0.06 | 0.14 | -0.18 |
| AvgDepsBl_nsubj | 0.03 | 0.9 | -0.1 | -0.1 | 0.08 |
| AvgDepsSen_nsubj | 0.94 | 0.14 | -0.07 | 0.01 | 0.14 |
| AvgDepsBl_nsubjpass | 0.05 | 0.2 | 0.03 | 0.08 | 0.16 |
| AvgDepsBl_nummod | -0.09 | 0.17 | -0.05 | 0.04 | -0.2 |
| AvgDepsBl_punct | -0.53 | 0.52 | -0.01 | -0.06 | -0.17 |
| AvgDepsSen_punct | -0.13 | 0.35 | 0.14 | 0.12 | 0.26 |
| AvgDepsBl_xcomp | 0.08 | 0.49 | -0.03 | 0.01 | -0.11 |
| AvgDepsSen_xcomp | 0.63 | 0.15 | 0.02 | -0.06 | -0.1 |
ReaderBench Model 1f
This model was trained on principal component scores for spring data in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -9.2262 | 0.1219 | 0.7713 | 0.1603 | -0.3706 | -0.0217 | 0.3129 | 0.0233 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
PC1 = scores on 1st principal component extracted, …
Note: Importance is unavailable for support vector machines when PCA-based pre-processing is used (so all values for svm are 0).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| PC2 | 31.31 | 56.99 | 35.09 | 0 | 11.67 | 23.86 | 35.8 | 21.83 |
| PC1 | 11.79 | 6.72 | 17.18 | 0 | 3.69 | 5.61 | 8.89 | 21.83 |
| PC4 | 10.38 | 7.69 | 9.11 | 0 | 8.29 | 5.24 | 16.85 | 12.66 |
| PC44 | 4.45 | 0.7 | 0.97 | 0 | 7.79 | 1.41 | 10.78 | 3.71 |
| PC9 | 4.36 | 2.62 | 4.85 | 0 | 7.31 | 1.81 | 0 | 10.92 |
| PC27 | 3.4 | 0.68 | 0.79 | 0 | 1.88 | 1.75 | 12.84 | 0 |
| PC14 | 3.37 | 1.1 | 3.45 | 0 | 6.67 | 1.95 | 0 | 7.21 |
| PC43 | 2.78 | 1.23 | 1.19 | 0 | 9.29 | 1.77 | 0 | 0 |
| PC6 | 2.51 | 0.56 | 3.24 | 0 | 3.33 | 1.2 | 0.08 | 9.17 |
| PC16 | 2.38 | 0.82 | 1.17 | 0 | 1.74 | 1.2 | 6.85 | 0 |
| PC15 | 2.31 | 1.41 | 2.25 | 0 | 4.31 | 1.89 | 0 | 10.92 |
| PC5 | 2.22 | 0.82 | 2.97 | 0 | 2.79 | 2.34 | 0.2 | 1.75 |
| PC28 | 1.47 | 0.78 | 1.17 | 0 | 3.69 | 1.94 | 0 | 0 |
| PC10 | 1.46 | 1.85 | 1.73 | 0 | 2.08 | 1.98 | 0 | 0 |
| PC11 | 1.33 | 1.77 | 1.55 | 0 | 1.91 | 2.26 | 0 | 0 |
| PC37 | 1.29 | 0.57 | 0.8 | 0 | 3.79 | 1.57 | 0 | 0 |
| PC12 | 1.19 | 0.4 | 1.5 | 0 | 1.87 | 2.16 | 0 | 0 |
| PC34 | 1.08 | 0.19 | 0.77 | 0 | 3.06 | 0.68 | 0 | 0 |
| PC31 | 1.05 | 0.52 | 0.79 | 0 | 2.74 | 2.09 | 0 | 0 |
| PC30 | 0.9 | 0.05 | 0.21 | 0 | 0 | 0.29 | 4.02 | 0 |
| PC19 | 0.84 | 1.4 | 0.92 | 0 | 1.29 | 0.05 | 0 | 0 |
| PC23 | 0.8 | 0.38 | 0.83 | 0 | 1.59 | 1.37 | 0 | 0 |
| PC3 | 0.77 | 0.92 | 0.36 | 0 | 0 | 2.84 | 2.68 | 0 |
| PC40 | 0.75 | 0.22 | 0.5 | 0 | 2.14 | 1.16 | 0 | 0 |
| PC21 | 0.74 | 0.02 | 0.7 | 0 | 0.87 | 1.03 | 1 | 0 |
| PC45 | 0.73 | 0.12 | 0.43 | 0 | 2.23 | 1.46 | 0 | 0 |
| PC25 | 0.65 | 0.57 | 0.67 | 0 | 1.22 | 1.65 | 0 | 0 |
| PC18 | 0.43 | 0.14 | 0.62 | 0 | 0.45 | 2.08 | 0 | 0 |
| PC20 | 0.38 | 0.49 | 0.55 | 0 | 0.3 | 1.36 | 0 | 0 |
| PC38 | 0.37 | 0.35 | 0.35 | 0 | 0.72 | 1.69 | 0 | 0 |
| PC35 | 0.37 | 0.16 | 0.4 | 0 | 0.7 | 0 | 0 | 0 |
| PC32 | 0.27 | 0.8 | 0.34 | 0 | 0.17 | 1.33 | 0 | 0 |
| PC17 | 0.24 | 0.08 | 0.47 | 0 | 0 | 1.41 | 0 | 0 |
| PC39 | 0.23 | 0.46 | 0.29 | 0 | 0.25 | 1.31 | 0 | 0 |
| PC13 | 0.23 | 1.06 | 0.32 | 0 | 0 | 2.26 | 0 | 0 |
| PC22 | 0.21 | 0.42 | 0.36 | 0 | 0 | 1.86 | 0 | 0 |
| PC41 | 0.2 | 0.42 | 0.27 | 0 | 0.16 | 1.77 | 0 | 0 |
| PC7 | 0.18 | 0.41 | 0.3 | 0 | 0 | 1.37 | 0 | 0 |
| PC26 | 0.17 | 1.46 | 0.12 | 0 | 0 | 2.05 | 0 | 0 |
| PC24 | 0.12 | 1.74 | 0 | 0 | 0 | 1.36 | 0 | 0 |
| PC42 | 0.1 | 0.32 | 0.15 | 0 | 0 | 1.77 | 0 | 0 |
| PC8 | 0.07 | 0.36 | 0.06 | 0 | 0 | 1.67 | 0 | 0 |
| PC33 | 0.06 | 0.12 | 0.08 | 0 | 0 | 1.34 | 0 | 0 |
| PC36 | 0.06 | 0 | 0.11 | 0 | 0 | 0.72 | 0 | 0 |
| PC29 | 0.02 | 0.13 | 0.02 | 0 | 0 | 2.08 | 0 | 0 |
Proportion of Variance by Varimax Rotated Component (RC)
Due to space limitations, loadings for only the first five principal components are displayed.
| RC1 | RC2 | RC3 | RC4 | RC5 | |
|---|---|---|---|---|---|
| SS loadings | 49.09 | 28.90 | 15.04 | 12.78 | 8.00 |
| Proportion Var | 0.24 | 0.14 | 0.07 | 0.06 | 0.04 |
| Cumulative Var | 0.24 | 0.39 | 0.46 | 0.53 | 0.57 |
| Proportion Explained | 0.43 | 0.25 | 0.13 | 0.11 | 0.07 |
| Cumulative Proportion | 0.43 | 0.69 | 0.82 | 0.93 | 1.00 |
Varimax Rotated Loadings
| Metric | RC1 | RC2 | RC3 | RC4 | RC5 |
|---|---|---|---|---|---|
| Sentences | -0.62 | 0.59 | -0.01 | -0.09 | 0.19 |
| Words | 0.13 | 0.87 | -0.07 | 0.37 | 0.19 |
| Content.words | 0.02 | 0.85 | 0.01 | 0.36 | 0.07 |
| RdbltyFlesch | -0.83 | -0.11 | -0.08 | 0.04 | 0.04 |
| RdbltyFog | 0.9 | 0.08 | 0.01 | -0.01 | 0 |
| RdbltyKincaid | 0.89 | 0.07 | 0.02 | 0 | -0.01 |
| RdbltyDaleChall | 0.47 | -0.12 | 0.41 | -0.23 | 0.28 |
| AvgBlLen | -0.01 | 0.88 | 0.03 | 0.29 | -0.02 |
| AvgCommaBl | -0.18 | 0.28 | -0.06 | -0.18 | 0.05 |
| AvgSenLen | 0.94 | 0.15 | 0.04 | 0 | -0.04 |
| AvgSenBl | -0.62 | 0.59 | -0.01 | -0.09 | 0.19 |
| AvgUnqWdBl | 0.02 | 0.89 | 0.06 | 0.22 | 0.02 |
| AvgUnqWdSen | 0.95 | 0.12 | 0.03 | 0.01 | -0.04 |
| AvgWdLen | -0.12 | 0.48 | 0.04 | -0.15 | -0.47 |
| AvgWdBl | 0.02 | 0.85 | 0.01 | 0.36 | 0.07 |
| AvgWdSen | 0.95 | 0.1 | 0.02 | 0.01 | 0.03 |
| CharEnt | -0.18 | 0.59 | -0.07 | -0.04 | 0.21 |
| SenStDevUnqWd | -0.35 | 0.23 | 0.05 | 0.67 | 0.07 |
| SenStdDevWd | -0.29 | 0.21 | 0.03 | 0.67 | 0.08 |
| WdEnt | -0.01 | 0.88 | 0 | 0.12 | 0.16 |
| WdLettStdDev | -0.04 | 0.56 | 0.08 | 0.02 | -0.16 |
| LxcDiv | 0.08 | 0.81 | 0.05 | 0.16 | -0.06 |
| LxcSoph | 0.56 | 0.08 | 0.09 | -0.08 | -0.48 |
| SynSoph | 0.89 | 0.25 | 0.04 | 0.2 | 0.03 |
| AvgNounBl | 0.13 | 0.55 | 0.22 | 0.28 | 0.52 |
| AvgPronounBl | 0.04 | 0.71 | -0.19 | 0.37 | 0.11 |
| AvgVerbBl | 0.04 | 0.84 | -0.17 | 0.36 | 0.02 |
| AvgAdverbBl | 0.2 | 0.61 | -0.01 | 0.25 | -0.12 |
| AvgAdjectiveBl | -0.08 | 0.65 | 0.05 | 0.06 | 0.04 |
| AvgPrepositionBl | 0.11 | 0.79 | -0.1 | 0.18 | 0.08 |
| AvgNounSen | 0.89 | -0.04 | 0.07 | -0.01 | 0.26 |
| AvgPronounSen | 0.9 | -0.01 | -0.07 | 0.03 | 0.02 |
| AvgVerbSen | 0.97 | 0.08 | -0.05 | 0 | -0.04 |
| AvgAdverbSen | 0.8 | 0.17 | 0.04 | 0.09 | -0.15 |
| AvgAdjectiveSen | 0.81 | 0 | 0 | -0.16 | -0.05 |
| AvgPrepositionSen | 0.9 | 0.17 | 0 | 0 | 0.02 |
| AvgUnqNoundBl | 0.13 | 0.52 | 0.2 | 0.16 | 0.48 |
| AvgUnqPronounBl | -0.01 | 0.54 | -0.15 | 0.18 | 0.09 |
| AvgUnqVerbBl | 0.01 | 0.8 | -0.09 | 0.29 | 0.01 |
| AvgUnqAdverbBl | 0.12 | 0.69 | -0.01 | 0.1 | -0.11 |
| AvgUnqAdjectiveBl | -0.07 | 0.65 | 0.04 | -0.02 | 0.04 |
| AvgUnqPrepositionBl | 0.04 | 0.77 | -0.11 | 0.17 | 0.05 |
| AvgPronBl_first_person | -0.02 | 0.47 | -0.22 | 0.38 | 0.16 |
| AvgPronBl_indefinite | -0.03 | 0.57 | -0.13 | -0.04 | 0.14 |
| AggPronSen_indefinite | 0.74 | 0.09 | -0.09 | -0.12 | 0.15 |
| AvgPronBl_third_person | 0.08 | 0.54 | -0.03 | 0.14 | -0.03 |
| AggPronSen_third_person | 0.78 | 0.06 | -0.02 | -0.05 | -0.13 |
| AvgSemDep | 0.98 | 0.06 | -0.01 | 0.01 | 0.08 |
| WdDiffLemmaStem | -0.11 | 0.34 | 0.02 | -0.16 | -0.19 |
| WdDiffWdStem | -0.11 | 0.39 | -0.03 | -0.1 | -0.42 |
| WdMaxDpthHypernymTree | 0.02 | -0.21 | 0.13 | 0.1 | 0.23 |
| WdAvgDpthHypernymTree | 0.02 | -0.22 | 0.09 | 0.1 | 0.19 |
| WdPathCntHypernymTree | 0.01 | -0.23 | 0.09 | 0.04 | 0.27 |
| WdPolysemyCnt | -0.04 | -0.04 | -0.4 | 0.25 | 0.07 |
| WdSylCnt | -0.11 | 0.5 | 0.32 | -0.08 | -0.3 |
| AvgAOADoc_Shock | -0.06 | 0.48 | -0.01 | 0.02 | -0.15 |
| AvgAOABl_Shock | -0.06 | 0.48 | -0.01 | 0.02 | -0.15 |
| AvgAOASen_Shock | 0.49 | 0.17 | -0.02 | 0.08 | -0.39 |
| AvgAOADoc_Cortese | -0.09 | -0.14 | 0.49 | -0.16 | -0.23 |
| AvgAOABl_Cortese | -0.09 | -0.14 | 0.49 | -0.16 | -0.23 |
| AvgAOASen_Cortese | 0.25 | -0.19 | 0.35 | 0.03 | -0.39 |
| AvgAOADoc_Kuperman | -0.1 | -0.04 | 0.64 | -0.1 | -0.12 |
| AvgAOABl_Kuperman | -0.1 | -0.04 | 0.64 | -0.1 | -0.12 |
| AvgAOASen_Kuperman | 0.11 | -0.05 | 0.62 | 0 | -0.32 |
| AvgAOADoc_Bird | -0.01 | 0.07 | 0.58 | 0.1 | -0.3 |
| AvgAOABl_Bird | -0.01 | 0.07 | 0.58 | 0.1 | -0.3 |
| AvgAOASen_Bird | 0.35 | -0.02 | 0.26 | 0.18 | -0.53 |
| AvgAOADoc_Bristol | -0.07 | 0.14 | 0.5 | 0.02 | -0.31 |
| AvgAOABl_Bristol | -0.07 | 0.14 | 0.5 | 0.02 | -0.31 |
| AvgAOASen_Bristol | 0.41 | -0.05 | 0.1 | 0.01 | -0.51 |
| AvgAOEDoc_IndexPolyFAT.3 | -0.04 | -0.1 | 0.89 | 0.02 | 0.22 |
| AvgAOEBl_IndexPolyFAT.3 | -0.04 | -0.1 | 0.89 | 0.02 | 0.22 |
| AvgAOESen_IndexPolyFAT.3 | 0.06 | -0.03 | 0.83 | 0.04 | 0.08 |
| AvgAOEDoc_InvLinRegSlo | -0.05 | -0.2 | 0.84 | -0.02 | 0.3 |
| AvgAOEBl_InvLinRegSlo | -0.05 | -0.2 | 0.84 | -0.02 | 0.3 |
| AvgAOESen_InvLinRegSlo | 0.17 | -0.15 | 0.69 | 0.07 | -0.03 |
| AvgAOEDoc_InfPointPoly | -0.03 | -0.05 | 0.85 | -0.11 | 0.22 |
| AvgAOEBl_InfPointPoly | -0.03 | -0.05 | 0.85 | -0.11 | 0.22 |
| AvgAOESen_InfPointPoly | 0.21 | -0.05 | 0.7 | 0.01 | -0.14 |
| AvgAOEDoc_InvAverage | -0.04 | -0.08 | 0.88 | -0.13 | 0.22 |
| AvgAOEBl_InvAverage | -0.04 | -0.08 | 0.88 | -0.13 | 0.22 |
| AvgAOESen_InvAverage | 0.2 | -0.06 | 0.73 | -0.01 | -0.13 |
| AvgAOEDoc_IndexAbThr.0.3. | -0.07 | -0.07 | 0.81 | 0.06 | 0.23 |
| AvgAOEBl_IndexAbThr.0.3. | -0.07 | -0.07 | 0.81 | 0.06 | 0.23 |
| AvgAOESen_IndexAbThr.0.3. | -0.03 | 0.02 | 0.78 | 0.04 | 0.18 |
| AvgNmdEntBl | 0.06 | 0.31 | 0.06 | 0.04 | 0.62 |
| AvgNounNmdEntBl | 0.04 | 0.16 | 0.2 | 0 | 0.67 |
| AvgUnqNmdEntBl | 0.03 | 0.34 | 0.06 | 0.03 | 0.63 |
| AvgNmdEntSen | 0.57 | -0.06 | 0.08 | 0 | 0.43 |
| TCorefChainDoc | 0.02 | 0.51 | -0.04 | 0.33 | 0.25 |
| AvgCorefChain | 0 | 0.44 | -0.14 | 0.18 | 0.03 |
| AvgChainSpan | 0.17 | 0.66 | -0.09 | 0.15 | -0.05 |
| AvgInferenceDistChain | 0.26 | 0.29 | -0.06 | 0.06 | 0.15 |
| TActCorefChainWd | -0.09 | -0.34 | -0.03 | -0.01 | 0.03 |
| TCorefChainBigSpan | 0.21 | 0.34 | -0.02 | 0.16 | 0.02 |
| AvgConnBl_addition | 0.45 | 0.25 | -0.03 | 0.6 | 0.11 |
| AvgConnSen_addition | 0.85 | -0.02 | 0.02 | 0.17 | 0.09 |
| AvgConnBl_conjunctions | 0.48 | 0.32 | -0.03 | 0.43 | 0.19 |
| AvgConnSen_conjunctions | 0.91 | -0.02 | 0.02 | 0.05 | 0.11 |
| AvgConnBl_contrasts | 0.16 | 0.51 | 0.12 | -0.15 | 0 |
| AvgConnSen_contrasts | 0.67 | 0.22 | 0.05 | -0.24 | -0.1 |
| AvgConnBl_coord_conjs | 0.11 | 0.28 | -0.06 | 0.38 | -0.23 |
| AvgConnSen_coord_conjs | 0.5 | 0.11 | 0.01 | 0.15 | -0.25 |
| AvgConnBl_coord_connects | 0.47 | 0.41 | -0.03 | 0.52 | 0.09 |
| AvgConnSen_coord_connects | 0.94 | 0.01 | 0.02 | 0.08 | 0.05 |
| AvgConnBl_logical_conns | 0.44 | 0.24 | -0.04 | 0.5 | 0.17 |
| AvgConnSen_logical_conns | 0.86 | -0.04 | 0.03 | 0.12 | 0.1 |
| AvgConnBl_oppositions | 0.17 | 0.49 | 0.07 | -0.22 | 0.04 |
| AvgConnSen_oppositions | 0.66 | 0.19 | 0.03 | -0.29 | -0.05 |
| AvgConnBl_order | 0.04 | 0.16 | -0.02 | 0.45 | -0.06 |
| AvgConnSen_order | 0.4 | -0.09 | 0.01 | 0.28 | 0.01 |
| AvgConnBl_reas_purp | 0.07 | 0.32 | -0.03 | 0.49 | -0.16 |
| AvgConnSen_reas_purp | 0.58 | 0.04 | 0.03 | 0.25 | -0.17 |
| AvgConnBl_semi_coords | 0.11 | 0.28 | -0.06 | 0.38 | -0.23 |
| AvgConnSen_semi_coords | 0.5 | 0.11 | 0.01 | 0.15 | -0.25 |
| AvgConnBl_sentence_link | 0.39 | 0.51 | -0.1 | 0.58 | 0.07 |
| AvgConnSen_sentence_link | 0.95 | 0 | 0 | 0.11 | 0.06 |
| AvgConnBl_simp_subords | -0.13 | 0.52 | -0.14 | -0.04 | 0.01 |
| AvgConnSen_simp_subords | 0.52 | 0.16 | -0.03 | -0.1 | -0.02 |
| AvgConnBl_temp_conns | -0.1 | 0.34 | -0.21 | 0.22 | 0.04 |
| AvgConnSen_temp_conns | 0.33 | 0 | -0.12 | 0.07 | 0.06 |
| LexChainAvgSpan | -0.04 | 0.29 | -0.21 | 0.47 | 0.17 |
| LexChainMaxSp | 0.04 | 0.61 | -0.1 | 0.45 | 0.02 |
| AvgBlScore | 0.22 | 0.56 | -0.18 | 0.53 | 0.08 |
| AvgSenScore | 0.93 | 0.05 | -0.06 | 0.06 | 0.07 |
| SenScoreStDev | -0.39 | 0.27 | -0.02 | 0.67 | 0.07 |
| AvgIntraBlCoh_LeackChod | -0.71 | 0.35 | 0.01 | 0.54 | 0.08 |
| AvgSenAdjCoh_LeackChod | -0.69 | 0.33 | 0.02 | 0.56 | 0.07 |
| AvgSenBlCoh_LeackChod | 0.76 | -0.5 | -0.07 | -0.05 | -0.17 |
| AvgIntraBlCoh_WuPalmer | -0.71 | 0.35 | 0.01 | 0.53 | 0.08 |
| AvgSenAdjCoh_WuPalmer | -0.7 | 0.33 | 0.01 | 0.55 | 0.08 |
| AvgSenBlCoh_WuPalmer | 0.76 | -0.51 | -0.08 | -0.07 | -0.17 |
| AvgIntraBlCoh_Path | -0.72 | 0.34 | 0 | 0.53 | 0.09 |
| AvgSenAdjCoh_Path | -0.7 | 0.32 | 0 | 0.55 | 0.08 |
| AvgSenBlCoh_Path | 0.78 | -0.52 | -0.07 | -0.17 | -0.14 |
| AvgIntraBlCoh_LSA | -0.7 | 0.35 | -0.01 | 0.55 | 0.09 |
| AvgSenAdjCoh_LSA | -0.68 | 0.34 | -0.01 | 0.57 | 0.09 |
| AvgSenBlCoh_LSA | 0.76 | -0.49 | -0.13 | -0.04 | -0.15 |
| AvgIntraBlCoh_LDA | -0.7 | 0.37 | -0.02 | 0.53 | 0.08 |
| AvgSenAdjCoh_LDA | -0.68 | 0.35 | -0.02 | 0.55 | 0.07 |
| AvgSenBlCoh_LDA | 0.71 | -0.39 | -0.2 | 0.02 | -0.19 |
| AvgIntraBlCoh_word2vec | -0.69 | 0.35 | 0.01 | 0.55 | 0.08 |
| AvgSenAdjCoh_word2vec | -0.69 | 0.33 | 0 | 0.56 | 0.09 |
| AvgSenBlCoh_word2vec | 0.76 | -0.53 | -0.09 | -0.07 | -0.13 |
| AvgBlVoiceCoOcc | -0.05 | 0.47 | -0.14 | 0.51 | 0.08 |
| AvgVoice | -0.05 | 0.46 | -0.15 | 0.54 | 0.08 |
| AvgSenSyll | 0.98 | 0.06 | 0.01 | -0.01 | 0.06 |
| AvgSenStressedSyll | 0.96 | 0.08 | 0.03 | 0.01 | 0.06 |
| AvgRhythmUnits | 0.18 | 0 | -0.08 | -0.18 | 0.01 |
| AvgRhythmUnitSyll | 0.81 | -0.04 | 0.02 | 0.09 | 0.09 |
| AvgRhythmUnitStreesSyll | 0.8 | -0.03 | 0.04 | 0.11 | 0.1 |
| LangRhythmCoeff | 0.27 | -0.25 | -0.03 | 0.24 | 0.22 |
| LangRhythmId | -0.19 | 0.11 | 0.02 | -0.04 | 0.21 |
| FrqRhythmId | 0.68 | -0.44 | -0.04 | -0.07 | -0.11 |
| LangRhythmDiameter | 0.34 | 0.07 | 0 | 0.11 | 0.35 |
| SenAsson | -0.05 | 0.28 | -0.05 | 0.17 | -0.09 |
| AvgDepsBl_acl | -0.14 | 0.18 | -0.12 | 0.02 | -0.13 |
| AvgDepsSen_acl | 0.12 | -0.23 | -0.17 | -0.14 | -0.17 |
| AvgDepsBl_advcl | 0.14 | 0.49 | -0.08 | 0.25 | -0.2 |
| AvgDepsSen_advcl | 0.66 | 0.18 | 0.02 | 0.03 | -0.23 |
| AvgDepsBl_advmod | 0.14 | 0.61 | -0.03 | 0.33 | -0.12 |
| AvgDepsSen_advmod | 0.79 | 0.15 | 0.01 | 0.15 | -0.12 |
| AvgDepsBl_amod | -0.1 | 0.58 | 0.09 | 0.11 | 0.15 |
| AvgDepsSen_amod | 0.7 | 0 | 0.02 | -0.07 | 0.09 |
| AvgDepsBl_aux | 0.16 | 0.54 | -0.15 | 0.06 | -0.03 |
| AvgDepsSen_aux | 0.76 | 0.21 | -0.09 | -0.06 | -0.08 |
| AvgDepsBl_auxpass | 0.04 | 0.4 | -0.05 | 0.2 | -0.08 |
| AvgDepsBl_case | 0.08 | 0.67 | -0.08 | 0.16 | 0.28 |
| AvgDepsSen_case | 0.84 | 0.1 | -0.03 | -0.01 | 0.17 |
| AvgDepsBl_cc | 0.49 | 0.35 | 0 | 0.43 | 0.19 |
| AvgDepsSen_cc | 0.92 | 0 | 0.03 | 0.05 | 0.09 |
| AvgDepsBl_ccomp | 0.2 | 0.28 | -0.25 | 0.31 | 0.11 |
| AvgDepsSen_ccomp | 0.76 | -0.05 | -0.15 | -0.02 | 0.11 |
| AvgDepsBl_compound | -0.02 | -0.06 | 0.5 | 0.1 | 0.4 |
| AvgDepsSen_compound | 0.37 | -0.22 | 0.24 | 0.02 | 0.28 |
| AvgDepsBl_conj | 0.6 | 0.31 | 0.02 | 0.37 | 0.15 |
| AvgDepsSen_conj | 0.88 | 0.02 | 0.02 | 0.03 | 0.08 |
| AvgDepsBl_cop | -0.05 | 0.49 | -0.09 | -0.11 | -0.03 |
| AvgDepsSen_cop | 0.64 | 0.02 | -0.13 | -0.16 | -0.15 |
| AvgDepsBl_dep | 0.38 | 0.28 | 0.03 | 0.25 | 0.25 |
| AvgDepsSen_dep | 0.73 | 0.01 | 0.05 | 0.02 | 0.18 |
| AvgDepsBl_det | 0.14 | 0.58 | 0.03 | 0.31 | 0.22 |
| AvgDepsSen_det | 0.76 | 0.11 | -0.01 | 0.03 | 0.19 |
| AvgDepsBl_dobj | 0.14 | 0.56 | -0.02 | 0.47 | 0.08 |
| AvgDepsSen_dobj | 0.9 | -0.03 | 0 | 0.06 | 0.06 |
| AvgDepsBl_mark | 0.09 | 0.6 | -0.1 | 0.25 | -0.23 |
| AvgDepsSen_mark | 0.75 | 0.1 | -0.03 | 0 | -0.24 |
| AvgDepsBl_mwe | 0.14 | 0.34 | 0.04 | 0.06 | 0.15 |
| AvgDepsSen_mwe | 0.45 | 0.15 | 0.08 | 0.04 | 0.14 |
| AvgDepsBl_neg | 0 | 0.38 | 0.01 | -0.11 | 0.02 |
| AvgDepsSen_neg | 0.39 | 0.05 | 0.05 | -0.21 | -0.07 |
| AvgDepsBl_nmod | 0.09 | 0.62 | -0.1 | 0.16 | 0.3 |
| AvgDepsSen_nmod | 0.83 | 0.08 | -0.03 | -0.03 | 0.2 |
| AvgDepsBl_nsubj | 0.07 | 0.79 | -0.16 | 0.34 | 0.15 |
| AvgDepsSen_nsubj | 0.97 | 0.03 | -0.04 | 0 | 0.04 |
| AvgDepsBl_nsubjpass | 0.05 | 0.34 | -0.04 | 0.2 | -0.04 |
| AvgDepsBl_nummod | 0.02 | 0.2 | -0.15 | 0.06 | 0.24 |
| AvgDepsBl_punct | -0.53 | 0.62 | -0.01 | -0.15 | 0.13 |
| AvgDepsSen_punct | -0.11 | 0.31 | 0 | -0.06 | -0.15 |
| AvgDepsBl_xcomp | 0.03 | 0.41 | 0.03 | 0.34 | -0.22 |
| AvgDepsSen_xcomp | 0.71 | 0.07 | 0.07 | 0.06 | -0.24 |
ReaderBench Model 2
General Description
ReaderBench Model 2 is a simplified version of Model 1 that better handles multi-paragraph compositions, and Model 2 is recommended over Model 1.
Model 2 is an ensemble (formed by averaging predicted quality scores) of three sub-models that are described below. All of these models used ReaderBench scores on 7 min narrative writing samples (“I once had a magic pencil and …”) from students in the fall, winter, and spring of Grades 2-5 (Mercer et al., 2019) to predict holistic writing quality on the samples (elo ratings calculated from paired comparisons). More details on the sample are available in (Mercer et al., 2019).
Highly correlated ReaderBench metrics (r > |.90|) were excluded during pre-processing (see section on Scoring Model Development for more details).
ReaderBench Model 2a
This model was trained with fall data from (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * pls = partial least squares regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * svm = support vector machines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | rf | mars | svm | cube |
|---|---|---|---|---|---|
| -4.338 | 0.2371 | 0.1755 | 0.1780 | 0.2234 | 0.2532 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | rf | mars | svm | cube |
|---|---|---|---|---|---|---|
| WdEnt | 20.53 | 4.67 | 10.12 | 73.84 | 5.16 | 18.67 |
| AvgDepsSen_dep | 4.65 | 1.23 | 0.88 | 16.82 | 0.88 | 5.25 |
| Content.words | 4.59 | 4.32 | 4.77 | 0 | 4.68 | 7.87 |
| Words | 3.72 | 4.44 | 4.67 | 0 | 4.67 | 4.17 |
| LxcDiv | 3.1 | 4.08 | 3.29 | 0 | 4.06 | 3.4 |
| AvgAOASen_Shock | 2.77 | 1.45 | 1.16 | 9.34 | 1.39 | 1.7 |
| TCorefChainDoc | 2.62 | 2.98 | 0.81 | 0 | 2.06 | 5.86 |
| AvgChainSpan | 2.59 | 3.27 | 3.83 | 0 | 3.1 | 2.47 |
| WdDiffWdStem | 2.46 | 2.73 | 3.07 | 0 | 2.24 | 3.7 |
| SynSoph | 2.12 | 1.71 | 0.92 | 0 | 1.82 | 5.09 |
| AvgDepsSen_punct | 2.03 | 2.48 | 1.68 | 0 | 1.74 | 3.55 |
| TActCorefChainWd | 1.93 | 1.6 | 1.91 | 0 | 1.47 | 4.01 |
| WdDiffLemmaStem | 1.66 | 1.52 | 0.72 | 0 | 2.44 | 2.93 |
| RdbltyFlesch | 1.55 | 0.77 | 1.22 | 0 | 1.09 | 4.01 |
| WdLettStdDev | 1.44 | 2.35 | 1.51 | 0 | 2.13 | 0.93 |
| AvgAOESen_InverseAverage | 1.37 | 1.44 | 1.22 | 0 | 1.09 | 2.62 |
| Sentences | 1.3 | 2.84 | 1.77 | 0 | 1.82 | 0 |
| AvgWdLen | 1.27 | 2.65 | 1.57 | 0 | 2.02 | 0 |
| LexChainMaxSp | 1.26 | 2.89 | 1.19 | 0 | 2.02 | 0 |
| AvgAOADoc_Shock | 1.26 | 2.36 | 1.89 | 0 | 1.68 | 0.31 |
| AvgAOADoc_Kuperman | 1.25 | 0.72 | 1.01 | 0 | 1.3 | 2.78 |
| WdSylCnt | 1.15 | 1.57 | 1.83 | 0 | 1.51 | 0.77 |
| CharEnt | 1.14 | 2.65 | 0.96 | 0 | 1.85 | 0 |
| LexChainAvgSpan | 1.12 | 2.18 | 1.5 | 0 | 1.86 | 0 |
| AvgDepsSen_advcl | 1.07 | 0.93 | 0.85 | 0 | 1.35 | 1.85 |
| AvgAOASen_Kuperman | 1.04 | 1.23 | 1.48 | 0 | 1.46 | 0.93 |
| AvgCorefChain | 1 | 1.86 | 0.85 | 0 | 0.9 | 1.08 |
| WdAvgDpthHypernymTree | 1 | 1.14 | 0.87 | 0 | 0.97 | 1.7 |
| SenStdDevWd | 0.98 | 1.96 | 1.43 | 0 | 1.49 | 0 |
| TCorefChainBigSpan | 0.95 | 2.16 | 1.44 | 0 | 1.13 | 0 |
| AvgAOADoc_Bristol | 0.94 | 1.75 | 1.03 | 0 | 1.1 | 0.62 |
| LxcSoph | 0.92 | 1.64 | 1.2 | 0 | 0.85 | 0.77 |
| AvgAdverbSen | 0.88 | 0.89 | 1.38 | 0 | 1.46 | 0.62 |
| RdbltyDaleChall | 0.87 | 1.75 | 1.63 | 0 | 1 | 0 |
| AvgSenAdjCoh_LDA | 0.82 | 1.97 | 0.64 | 0 | 1.33 | 0 |
| AvgRhythmUnits | 0.82 | 1.12 | 1.13 | 0 | 1.15 | 0.62 |
| FrqRhythmId | 0.8 | 1.69 | 1.07 | 0 | 1.18 | 0 |
| AvgAOADoc_Bird | 0.78 | 0.95 | 0.3 | 0 | 1.43 | 0.93 |
| AvgVoice | 0.78 | 2.01 | 0.76 | 0 | 0.99 | 0 |
| AvgAOADoc_Cortese | 0.77 | 0.69 | 1.3 | 0 | 1.57 | 0.31 |
| WdPathCntHypernymTree | 0.71 | 1.45 | 0.84 | 0 | 1.17 | 0 |
| AvgConnSen_simple_subordinators | 0.7 | 0.51 | 2.49 | 0 | 0.82 | 0 |
| AvgAOASen_Bristol | 0.68 | 0.66 | 0.71 | 0 | 1.29 | 0.62 |
| AvgRhythmUnitStreesSyll | 0.63 | 0.08 | 0.91 | 0 | 0.81 | 1.23 |
| AvgInferenceDistChain | 0.62 | 1.39 | 0.34 | 0 | 1.2 | 0 |
| AggPronSen_indefinite | 0.62 | 0.45 | 0.63 | 0 | 1.31 | 0.62 |
| AvgAOASen_Bird | 0.6 | 1.13 | 0.37 | 0 | 1.37 | 0 |
| AvgDepsSen_compound | 0.6 | 0.72 | 0.5 | 0 | 0.48 | 1.08 |
| WdPolysemyCnt | 0.58 | 0 | 1.09 | 0 | 1.9 | 0 |
| AvgDepsSen_ccomp | 0.57 | 0.09 | 1.32 | 0 | 0.9 | 0.62 |
| AvgAOASen_Cortese | 0.55 | 1.15 | 0.3 | 0 | 1.17 | 0 |
| AvgDepsSen_cop | 0.54 | 0.24 | 0.58 | 0 | 0.97 | 0.77 |
| AvgPronounSen | 0.54 | 0.12 | 0.93 | 0 | 0.48 | 1.08 |
| AvgNmdEntSen | 0.52 | 0.24 | 1.12 | 0 | 1.33 | 0 |
| AvgNounSen | 0.52 | 0.24 | 0.15 | 0 | 0.18 | 1.7 |
| AvgDepsSen_nmod | 0.48 | 0.7 | 0.69 | 0 | 1 | 0 |
| AvgDepsSen_aux | 0.48 | 0.24 | 0.92 | 0 | 1.31 | 0 |
| AvgConnSen_addition | 0.48 | 1.1 | 0.6 | 0 | 0.66 | 0 |
| AvgDepsSen_dobj | 0.48 | 0.23 | 1.51 | 0 | 0.16 | 0.62 |
| AvgAOEDoc_InverseLinearRegressionSlope | 0.44 | 0.4 | 0.8 | 0 | 0.68 | 0.31 |
| AvgDepsSen_mark | 0.41 | 0.43 | 0.95 | 0 | 0.73 | 0 |
| AvgConnSen_temporal_connectors | 0.41 | 0.32 | 0.64 | 0 | 1.11 | 0 |
| AvgDepsSen_det | 0.4 | 0.18 | 0.4 | 0 | 0.72 | 0.62 |
| AvgConnSen_semi_coordinators | 0.38 | 0.8 | 0.15 | 0 | 0.16 | 0.62 |
| AvgConnSen_order | 0.36 | 0.31 | 1.74 | 0 | 0.03 | 0 |
| AggPronSen_third_person | 0.36 | 0.57 | 0.91 | 0 | 0.41 | 0 |
| LangRhythmDiameter | 0.35 | 0.57 | 0.79 | 0 | 0.08 | 0.31 |
| SenAsson | 0.35 | 0.8 | 0.83 | 0 | 0.16 | 0 |
| AvgAOEDoc_IndexAboveThreshold.0.3. | 0.33 | 0.03 | 0.43 | 0 | 0.87 | 0.31 |
| AvgDepsSen_amod | 0.29 | 0.33 | 0.98 | 0 | 0.27 | 0 |
| AvgAdjectiveSen | 0.28 | 0.1 | 1.28 | 0 | 0.21 | 0 |
| AvgConnSen_oppositions | 0.27 | 0.54 | 0.82 | 0 | 0.07 | 0 |
| AvgDepsSen_xcomp | 0.24 | 0.01 | 0.13 | 0 | 1.04 | 0 |
| AvgAOEDoc_IndexPolynomialFitAboveThreshold.0.3. | 0.21 | 0.12 | 0.1 | 0 | 0.78 | 0 |
| LangRhythmId | 0.19 | 0.47 | 0.45 | 0 | 0.05 | 0 |
| AvgDepsSen_neg | 0.18 | 0.03 | 1.05 | 0 | 0 | 0 |
| AvgDepsSen_mwe | 0.17 | 0.38 | 0.47 | 0 | 0.04 | 0 |
| LangRhythmCoeff | 0.16 | 0 | 0.22 | 0 | 0.61 | 0 |
| AvgDepsSen_acl | 0.06 | 0.25 | 0 | 0 | 0.02 | 0 |
ReaderBench Model 2b
This model was trained with winter data from (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * pls = partial least squares regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * svm = support vector machines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | rf | mars | svm | cube |
|---|---|---|---|---|---|
| -5.4658 | 0.2205 | 0.5768 | 0.2047 | 0.0528 | 0.0400 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | rf | mars | svm | cube |
|---|---|---|---|---|---|---|
| Content.words | 11.94 | 5.23 | 4.51 | 41.33 | 4.49 | 15.7 |
| WdEnt | 8.27 | 5.15 | 5.03 | 19.27 | 4.44 | 21.01 |
| SynSoph | 4.17 | 1.03 | 2.06 | 14.97 | 1.69 | 0 |
| LxcDiv | 3.24 | 4.93 | 3.5 | 0 | 3.86 | 5.8 |
| AvgDepsSen_det | 3.18 | 0.25 | 1.12 | 12.65 | 0.94 | 3.38 |
| TCorefChainDoc | 2.63 | 4 | 2.76 | 0 | 2.45 | 6.76 |
| AvgChainSpan | 2.28 | 3.64 | 2.57 | 0 | 2.88 | 1.45 |
| LexChainMaxSp | 2.25 | 3.56 | 2.72 | 0 | 1.98 | 0 |
| TActCorefChainWd | 2.22 | 0.83 | 0.71 | 7.46 | 0.78 | 6.76 |
| Sentences | 2.18 | 3.8 | 2.48 | 0 | 2.25 | 0 |
| AvgNounSen | 2.07 | 0.89 | 1.54 | 4.33 | 0.64 | 6.52 |
| CharEnt | 1.7 | 3.41 | 1.66 | 0 | 2.16 | 0.97 |
| RdbltyFlesch | 1.36 | 0.46 | 1.91 | 0 | 1.18 | 5.56 |
| WdLettStdDev | 1.31 | 2.58 | 1.31 | 0 | 2 | 0 |
| AvgSenAdjCoh_LeackockChodorow | 1.3 | 2.75 | 1.24 | 0 | 1.87 | 0 |
| FrqRhythmId | 1.28 | 2.48 | 1.39 | 0 | 1.13 | 0 |
| AvgDepsSen_aux | 1.25 | 0.94 | 1.69 | 0 | 0.97 | 3.38 |
| AvgWdLen | 1.24 | 2.36 | 1.26 | 0 | 2.04 | 0 |
| AvgAOADoc_Bristol | 1.21 | 1.59 | 1.61 | 0 | 0.88 | 0 |
| AvgDepsSen_compound | 1.2 | 1.17 | 1.78 | 0 | 0.48 | 0 |
| AvgVoice | 1.2 | 2.75 | 1.13 | 0 | 1.19 | 0 |
| AvgAOADoc_Shock | 1.16 | 2.54 | 1.13 | 0 | 1.17 | 0 |
| TCorefChainBigSpan | 1.13 | 2.24 | 1.22 | 0 | 0.78 | 0 |
| AvgConnSen_addition | 1.07 | 1.31 | 1.29 | 0 | 1.31 | 1.69 |
| WdDiffWdStem | 1.04 | 2.05 | 1.06 | 0 | 1.35 | 0 |
| AvgConnSen_logical_connectors | 1.03 | 1.49 | 1.11 | 0 | 1.27 | 2.17 |
| AvgCorefChain | 1.01 | 2.38 | 0.9 | 0 | 1.32 | 0 |
| AggPronSen_third_person | 0.98 | 1.18 | 1.23 | 0 | 0.56 | 1.93 |
| AvgDepsSen_punct | 0.98 | 1.89 | 1.01 | 0 | 1.48 | 0 |
| AvgDepsSen_dep | 0.95 | 1.01 | 1.31 | 0 | 1.07 | 0 |
| AvgRhythmUnitStreesSyll | 0.95 | 0.44 | 1.46 | 0 | 0.9 | 1.21 |
| AvgDepsSen_dobj | 0.95 | 1.12 | 1.04 | 0 | 1.07 | 3.38 |
| AvgAdjectiveSen | 0.91 | 0.44 | 1.47 | 0 | 0.9 | 0 |
| SenStdDevWd | 0.9 | 2.04 | 0.69 | 0 | 1.52 | 1.21 |
| LexChainAvgSpan | 0.87 | 1.85 | 0.77 | 0 | 1.88 | 0 |
| WdPathCntHypernymTree | 0.86 | 1.46 | 0.94 | 0 | 1.38 | 0 |
| AvgAOESen_InverseAverage | 0.85 | 0.71 | 1.27 | 0 | 0.81 | 0 |
| AvgDepsSen_mark | 0.83 | 0.22 | 1.4 | 0 | 1.06 | 0 |
| WdPolysemyCnt | 0.83 | 0.32 | 1.43 | 0 | 0.39 | 0 |
| AvgConnSen_reason_and_purpose | 0.82 | 0.35 | 1.3 | 0 | 0.7 | 0.97 |
| LangRhythmCoeff | 0.8 | 1.15 | 1 | 0 | 0.92 | 0 |
| AvgConnSen_simple_subordinators | 0.78 | 0.11 | 1.3 | 0 | 1.47 | 0 |
| AvgDepsSen_xcomp | 0.76 | 0.11 | 1.25 | 0 | 1.6 | 0 |
| AvgAOASen_Bird | 0.76 | 0.33 | 1.19 | 0 | 0.62 | 0.97 |
| AvgDepsSen_ccomp | 0.75 | 0.16 | 1.29 | 0 | 0.79 | 0 |
| RdbltyDaleChall | 0.75 | 2.41 | 0.41 | 0 | 1.07 | 0 |
| AvgAOEDoc_InflectionPointPolynomial | 0.73 | 0.7 | 1.06 | 0 | 0.65 | 0 |
| AvgAOESen_IndexAboveThreshold.0.3. | 0.7 | 0.47 | 1.01 | 0 | 1.43 | 0 |
| AvgAOESen_IndexPolynomialFitAboveThreshold.0.3. | 0.7 | 0.55 | 1.01 | 0 | 1.13 | 0 |
| AggPronSen_indefinite | 0.7 | 0.09 | 1.07 | 0 | 1.64 | 1.21 |
| AvgDepsSen_cop | 0.7 | 0.09 | 1.16 | 0 | 1.49 | 0 |
| AvgNmdEntSen | 0.68 | 0.45 | 1.02 | 0 | 1.07 | 0 |
| AvgConnSen_contrasts | 0.68 | 0.32 | 1.03 | 0 | 0.59 | 1.21 |
| AvgConnSen_oppositions | 0.68 | 0.07 | 1.18 | 0 | 0.94 | 0 |
| AvgDepsSen_advcl | 0.67 | 0.03 | 1.13 | 0 | 1.31 | 0 |
| AvgAdverbSen | 0.67 | 0.43 | 1.01 | 0 | 1.08 | 0 |
| AvgAOEDoc_IndexPolynomialFitAboveThreshold.0.3. | 0.66 | 0 | 1.14 | 0 | 1.18 | 0 |
| AvgDepsSen_nmod | 0.66 | 0.74 | 0.76 | 0 | 1.35 | 1.21 |
| AvgAOADoc_Bird | 0.65 | 0.95 | 0.77 | 0 | 1.11 | 0 |
| AvgDepsSen_amod | 0.65 | 0.53 | 0.69 | 0 | 0.9 | 3.86 |
| AvgConnSen_semi_coordinators | 0.64 | 0.24 | 1.04 | 0 | 0.78 | 0 |
| WdMaxDpthHypernymTree | 0.62 | 1.46 | 0.46 | 0 | 1.61 | 0 |
| AvgAOASen_Shock | 0.62 | 1.13 | 0.62 | 0 | 1.34 | 0 |
| AvgAOASen_Kuperman | 0.6 | 0.15 | 1.01 | 0 | 0.45 | 0.48 |
| AvgConnSen_temporal_connectors | 0.58 | 0.27 | 0.99 | 0 | 0.01 | 0 |
| AvgAOASen_Bristol | 0.57 | 0.38 | 0.87 | 0 | 0.67 | 0 |
| LangRhythmDiameter | 0.56 | 0.65 | 0.81 | 0 | 0.06 | 0 |
| AvgConnSen_order | 0.52 | 0.29 | 0.7 | 0 | 1 | 1.21 |
| AvgAOEDoc_IndexAboveThreshold.0.3. | 0.5 | 0.01 | 0.79 | 0 | 1.65 | 0 |
| AvgRhythmUnits | 0.5 | 0.73 | 0.57 | 0 | 1.13 | 0 |
| AvgAOADoc_Kuperman | 0.5 | 0.14 | 0.82 | 0 | 0.86 | 0 |
| AvgAOASen_Cortese | 0.49 | 0.13 | 0.83 | 0 | 0.66 | 0 |
| AvgInferenceDistChain | 0.48 | 0.87 | 0.51 | 0 | 0.71 | 0 |
| WdDiffLemmaStem | 0.48 | 0.4 | 0.62 | 0 | 1.55 | 0 |
| SenAsson | 0.42 | 0.99 | 0.4 | 0 | 0.15 | 0 |
| AvgDepsSen_mwe | 0.41 | 0.66 | 0.52 | 0 | 0.07 | 0 |
| AvgDepsSen_neg | 0.39 | 0.28 | 0.64 | 0 | 0 | 0 |
| AvgDepsSen_acl | 0.33 | 0.45 | 0.46 | 0 | 0.03 | 0 |
| LxcSoph | 0.31 | 0.39 | 0.35 | 0 | 0.92 | 0 |
| AvgAOEDoc_InverseLinearRegressionSlope | 0.27 | 0.19 | 0.39 | 0 | 0.61 | 0 |
| AvgAOADoc_Cortese | 0.24 | 0.76 | 0.09 | 0 | 0.85 | 0 |
| WdSylCnt | 0.23 | 0.83 | 0 | 0 | 1.29 | 0 |
| LangRhythmId | 0.03 | 0.09 | 0.03 | 0 | 0 | 0 |
ReaderBench Model 2c
This model was trained on spring data from (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * pls = partial least squares regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * gbm = stochastic gradient boosted trees * svm = support vector machines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | rf | mars | gbm | svm | cube |
|---|---|---|---|---|---|---|
| -7.3027 | 0.2354 | 0.1868 | 0.1595 | 0.1816 | 0.2191 | 0.0704 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | rf | mars | gbm | svm | cube |
|---|---|---|---|---|---|---|---|
| Content.words | 11.99 | 4.55 | 5.81 | 30.16 | 21.71 | 4.24 | 11.11 |
| WdEnt | 7.28 | 4.3 | 5.74 | 0 | 21.09 | 4.12 | 12.09 |
| AvgDepsSen_compound | 3.97 | 2.07 | 1.98 | 13.22 | 2.22 | 1.52 | 6.82 |
| AvgWdLen | 3.87 | 2.64 | 2.65 | 7.11 | 4.85 | 2.04 | 7.02 |
| LxcDiv | 3.77 | 4.06 | 4.13 | 0 | 7.72 | 3.59 | 0.78 |
| AvgChainSpan | 3.36 | 3.09 | 2.64 | 5.1 | 4.15 | 2.66 | 2.34 |
| TCorefChainBigSpan | 2.64 | 2.23 | 1.48 | 10.59 | 0.43 | 0.93 | 0 |
| Sentences | 2.37 | 3.33 | 2.15 | 0 | 2.63 | 2.24 | 4.87 |
| AvgDepsSen_mark | 2.21 | 0.38 | 1.17 | 10.59 | 0.08 | 1.45 | 0 |
| AvgDepsSen_dobj | 2 | 0.81 | 0.96 | 8.72 | 0.14 | 1.13 | 0.97 |
| AvgSenAdjCoh_LSA | 1.95 | 2.68 | 1.87 | 0 | 3.17 | 2.26 | 0 |
| AvgCorefChain | 1.94 | 2.2 | 1 | 5.1 | 0.28 | 1.28 | 2.73 |
| WdDiffWdStem | 1.92 | 2.4 | 1.86 | 0 | 2.95 | 2.09 | 1.56 |
| LexChainMaxSp | 1.82 | 3.13 | 2.35 | 0 | 1.28 | 2.01 | 0.97 |
| WdLettStdDev | 1.79 | 3 | 1.66 | 0 | 1.64 | 2.28 | 0.97 |
| TCorefChainDoc | 1.62 | 3.23 | 1.85 | 0 | 0.17 | 1.92 | 2.14 |
| CharEnt | 1.59 | 2.56 | 0.9 | 0 | 0.29 | 2.1 | 5.46 |
| WdSylCnt | 1.53 | 2.45 | 1.7 | 0 | 1.52 | 1.55 | 1.36 |
| FrqRhythmId | 1.47 | 2.67 | 1.7 | 0 | 1.03 | 1.59 | 0.97 |
| AvgDepsSen_punct | 1.36 | 1.82 | 1.57 | 0 | 0.72 | 1.83 | 2.53 |
| AvgAOEDoc_InverseLinearRegressionSlope | 1.32 | 1.31 | 0.73 | 4.26 | 0.24 | 0.89 | 0.39 |
| RdbltyDaleChall | 1.25 | 1.81 | 1.27 | 0 | 1.04 | 1.02 | 3.51 |
| AvgAOADoc_Shock | 1.2 | 2.2 | 1.24 | 0 | 0.69 | 1.8 | 0 |
| LangRhythmCoeff | 1.06 | 1.61 | 1.41 | 0 | 1.03 | 1.24 | 0.19 |
| LexChainAvgSpan | 1.05 | 1.94 | 1.36 | 0 | 0.16 | 1.66 | 0 |
| SenAsson | 1.05 | 1.63 | 0.58 | 3.07 | 0.02 | 0.56 | 0 |
| AvgVoice | 1 | 2.62 | 0.58 | 0 | 0 | 1.36 | 0.39 |
| AvgNounSen | 0.97 | 1.09 | 1.59 | 0 | 0.47 | 1.06 | 2.14 |
| WdDiffLemmaStem | 0.94 | 1.65 | 1.01 | 0 | 0.36 | 1.32 | 0.78 |
| TActCorefChainWd | 0.94 | 0.93 | 0.74 | 0 | 1.05 | 0.71 | 4.09 |
| AvgAOADoc_Cortese | 0.93 | 1.16 | 0.9 | 0 | 0.56 | 1.89 | 0.39 |
| AvgAOASen_Bristol | 0.92 | 0.35 | 1.28 | 2.08 | 1.15 | 0.34 | 0.39 |
| SenStdDevWd | 0.92 | 1.6 | 0.97 | 0 | 0.09 | 1.78 | 0 |
| AvgDepsSen_xcomp | 0.83 | 0.41 | 1.61 | 0 | 1.42 | 0.99 | 0 |
| AvgAdjectiveSen | 0.83 | 1.24 | 1 | 0 | 0.18 | 1.21 | 1.36 |
| AvgDepsSen_nmod | 0.81 | 0.16 | 1.23 | 0 | 0.38 | 1.25 | 3.51 |
| AvgAOADoc_Kuperman | 0.8 | 0.7 | 1.18 | 0 | 0.65 | 1.44 | 0.39 |
| AvgDepsSen_amod | 0.79 | 1.11 | 0.91 | 0 | 0.1 | 1.33 | 1.36 |
| AvgDepsSen_ccomp | 0.78 | 1.06 | 1.41 | 0 | 0.27 | 1.18 | 0 |
| AvgAOASen_Kuperman | 0.78 | 0.78 | 0.83 | 0 | 0.41 | 0.99 | 2.73 |
| AvgNmdEntSen | 0.78 | 0.93 | 1 | 0 | 1.05 | 1.05 | 0 |
| AvgAOESen_IndexPolynomialFitAboveThreshold.0.3. | 0.76 | 0.58 | 1.12 | 0 | 0.4 | 0.84 | 2.73 |
| AvgConnSen_simple_subordinators | 0.74 | 0.25 | 0.86 | 0 | 1.12 | 1.52 | 0.39 |
| AvgPronounSen | 0.72 | 1.09 | 1.13 | 0 | 0.02 | 0.99 | 0.97 |
| AvgAOASen_Shock | 0.69 | 0.48 | 1.51 | 0 | 0.21 | 1.32 | 0 |
| AvgConnSen_reason_and_purpose | 0.68 | 0.16 | 1.31 | 0 | 0.82 | 1.29 | 0 |
| AvgAOASen_Cortese | 0.66 | 1.25 | 0.45 | 0 | 0.24 | 1.22 | 0 |
| AvgAOESen_InverseLinearRegressionSlope | 0.66 | 0.99 | 1.02 | 0 | 0.31 | 0.68 | 0.97 |
| AvgAOEDoc_InflectionPointPolynomial | 0.65 | 0.64 | 0.36 | 0 | 0.37 | 0.61 | 3.7 |
| AvgConnSen_addition | 0.65 | 0.88 | 0.88 | 0 | 0.12 | 1.21 | 0.39 |
| AvgConnSen_order | 0.64 | 0.44 | 0.48 | 0 | 0.92 | 1.41 | 0 |
| AvgInferenceDistChain | 0.64 | 0.8 | 0.91 | 0 | 0.7 | 0.83 | 0 |
| WdPolysemyCnt | 0.62 | 0.27 | 0.93 | 0 | 0.37 | 1.61 | 0 |
| AvgAOEDoc_IndexPolynomialFitAboveThreshold.0.3. | 0.61 | 0.83 | 0.71 | 0 | 0.07 | 1.07 | 0.97 |
| AvgRhythmUnits | 0.61 | 0.3 | 1.24 | 0 | 0.32 | 1.3 | 0 |
| AvgDepsSen_aux | 0.57 | 0.03 | 1.26 | 0 | 0.38 | 1.32 | 0 |
| SynSoph | 0.57 | 0.59 | 0.85 | 0 | 0.08 | 1.02 | 0.97 |
| AvgDepsSen_cop | 0.55 | 0.87 | 0.48 | 0 | 0.05 | 1.25 | 0 |
| AvgRhythmUnitStreesSyll | 0.52 | 0.76 | 1.17 | 0 | 0.14 | 0.56 | 0 |
| AvgDepsSen_advmod | 0.48 | 0.33 | 0.6 | 0 | 0.2 | 1.26 | 0 |
| AvgDepsSen_det | 0.48 | 0.22 | 1.04 | 0 | 0.45 | 0.68 | 0.39 |
| AggPronSen_third_person | 0.47 | 0.86 | 0.8 | 0 | 0.08 | 0.58 | 0 |
| AvgAOADoc_Bristol | 0.45 | 0.36 | 0.71 | 0 | 0.12 | 1.02 | 0.19 |
| AvgDepsSen_acl | 0.44 | 1.28 | 0.29 | 0 | 0.16 | 0.36 | 0 |
| AvgAOADoc_Bird | 0.44 | 0.38 | 0.84 | 0 | 0.13 | 0.89 | 0 |
| WdAvgDpthHypernymTree | 0.43 | 0.79 | 0.71 | 0 | 0.06 | 0.54 | 0 |
| RdbltyFlesch | 0.43 | 0.42 | 1.35 | 0 | 0.03 | 0.44 | 0 |
| AvgDepsSen_dep | 0.42 | 0.68 | 0.6 | 0 | 0.02 | 0.75 | 0 |
| AggPronSen_indefinite | 0.41 | 0.34 | 0.51 | 0 | 0.14 | 1.05 | 0 |
| AvgConnSen_semi_coordinators | 0.39 | 0 | 1.01 | 0 | 0.13 | 0.92 | 0 |
| AvgDepsSen_mwe | 0.38 | 0.6 | 1.17 | 0 | 0.1 | 0.07 | 0 |
| AvgDepsSen_advcl | 0.38 | 0.06 | 0.44 | 0 | 0.01 | 1.4 | 0 |
| AvgDepsSen_neg | 0.37 | 0.51 | 0.97 | 0 | 0.4 | 0.05 | 0 |
| WdPathCntHypernymTree | 0.36 | 0.89 | 0.33 | 0 | 0.19 | 0.35 | 0 |
| AvgAOESen_IndexAboveThreshold.0.3. | 0.35 | 0.27 | 0 | 0 | 0.41 | 1.05 | 0 |
| AvgAOASen_Bird | 0.33 | 0.04 | 0.75 | 0 | 0.59 | 0.42 | 0 |
| LxcSoph | 0.31 | 0.02 | 0.61 | 0 | 0.16 | 0.18 | 1.95 |
| AvgConnSen_oppositions | 0.26 | 0.11 | 0.98 | 0 | 0.36 | 0 | 0 |
| LangRhythmDiameter | 0.24 | 0.3 | 0.84 | 0 | 0.12 | 0.03 | 0 |
| AvgConnSen_temporal_connectors | 0.17 | 0.23 | 0.58 | 0 | 0.09 | 0.01 | 0 |
| LangRhythmId | 0.09 | 0.22 | 0.23 | 0 | 0.02 | 0.01 | 0 |
ReaderBench Model 3
General Description
ReaderBench Model 3, recommended for current use, is an ensemble (formed by averaging predicted quality scores) of three genre-specific models, detailed below.
The models were trained on ReaderBench scores from 15 min narrative, expository, and persuasive writing samples from students in Grades 2-5 to predict holistic writing quality on the samples (theta scores calculated from paired comparisons).
Highly correlated ReaderBench metrics (r > |.90|) were excluded during pre-processing (see section on Scoring Model Development for more details).
More details on the sample will be provided once peer review is complete on the main study using this model.
ReaderBench Model 3narr
This model was trained on 15-minute narrative writing samples.
Algorithm Weightings in Ensemble
Abbreviations: * overall = ensemble model * pls = partial least squares regression * gbm = stochastic gradient boosted trees * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | rf | mars | gbm | svm | enet | cube |
|---|---|---|---|---|---|---|---|
| 0.0000 | 0.1419 | 0.0945 | 0.3143 | 0.0729 | 0.0816 | 0.1792 | 0.1538 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | rf | mars | gbm | svm | enet | cube |
|---|---|---|---|---|---|---|---|---|
| Content.words | 13.76 | 2 | 2.22 | 32.41 | 16.24 | 2.38 | 4.95 | 8.73 |
| RB.AvgWdLen | 7.5 | 0.81 | 1.06 | 21.05 | 1.14 | 1.01 | 1.38 | 3.55 |
| RB.AvgDepsBl_compound | 4.66 | 0.89 | 1.01 | 11.15 | 2.17 | 0.45 | 2.45 | 3.09 |
| RB.WdEnt | 4.59 | 1.7 | 2.06 | 7.03 | 4.52 | 1.75 | 4.27 | 5.73 |
| RB.LangRhythmId | 3.17 | 0.69 | 0.35 | 8.26 | 0.06 | 0.21 | 2.85 | 0.18 |
| RB.RdbltyDaleChall | 3.15 | 1.27 | 1.24 | 4.63 | 2.86 | 0.95 | 4.05 | 3.27 |
| RB.AvgUnqWdBl | 2.75 | 1.63 | 1.92 | 4.02 | 0.46 | 1.82 | 0 | 6.45 |
| RB.LxcDiv | 2.67 | 1.87 | 1.89 | 0 | 11.82 | 2.04 | 3.58 | 4.27 |
| Sentences | 2.62 | 1.71 | 2.13 | 0 | 5.6 | 1.52 | 4.67 | 5.91 |
| RB.TCorefChainDoc | 2.35 | 1.99 | 1.91 | 0 | 5.33 | 1.97 | 5.29 | 3.09 |
| RB.AvgAOADoc_Cortese | 1.8 | 0.21 | 0.37 | 3.53 | 0.76 | 0.8 | 2.06 | 1.36 |
| RB.CAF | 1.78 | 1.83 | 1.52 | 0 | 1.84 | 1.87 | 3.65 | 3.27 |
| RB.AvgNounNmdEntBl | 1.6 | 0.53 | 0.45 | 4.63 | 0.21 | 0.19 | 0 | 0.36 |
| RB.AvgDepsBl_nsubjpass | 1.42 | 0.81 | 0.27 | 3.29 | 0.2 | 0.41 | 1.23 | 0.18 |
| RB.AvgDepsBl_aux | 1.21 | 1.48 | 1.75 | 0 | 2.12 | 1.53 | 1.3 | 2.36 |
| RB.TActCorefChainWd | 1.06 | 0.33 | 0.95 | 0 | 1.19 | 1.17 | 2.65 | 2 |
| RB.AvgDepsBl_nsubj | 1.05 | 1.67 | 1.78 | 0 | 2.94 | 1.85 | 0 | 2.09 |
| RB.AvgPronounBl | 0.99 | 1.72 | 1.84 | 0 | 3.59 | 2.06 | 0 | 1.18 |
| RB.AvgUnqNoundBl | 0.93 | 0.71 | 0.59 | 0 | 0.37 | 0.65 | 2.75 | 1.55 |
| RB.TCorefChainBigSpan | 0.89 | 1.61 | 1.19 | 0 | 0.16 | 1.33 | 2.58 | 0 |
| RB.AvgAOESen_InflectionPointPolynomial | 0.87 | 0.97 | 1.18 | 0 | 0.57 | 1.09 | 2.33 | 0.73 |
| RB.AvgBlScore | 0.83 | 1.46 | 1.28 | 0 | 0.92 | 1.62 | 0 | 2.18 |
| RB.AvgConnBl_addition | 0.81 | 0.99 | 0.83 | 0 | 0.77 | 0.66 | 1.36 | 1.73 |
| RB.AvgChainSpan | 0.81 | 1.52 | 1.28 | 0 | 2.39 | 1.7 | 0 | 1.27 |
| RB.AvgPrepositionBl | 0.79 | 1.51 | 1.03 | 0 | 0.76 | 1.6 | 0.95 | 1 |
| RB.AvgUnqPrepositionBl | 0.76 | 1.48 | 1.09 | 0 | 0.48 | 1.55 | 0.8 | 1.09 |
| RB.SenStdDevWd | 0.74 | 0.86 | 1.25 | 0 | 1.41 | 1.28 | 1.45 | 0.36 |
| RB.AvgAOADoc_Shock | 0.72 | 0.86 | 0.94 | 0 | 1.36 | 1.15 | 0.8 | 1.27 |
| RB.AvgDepsBl_punct | 0.68 | 1.26 | 1.34 | 0 | 1 | 1 | 0.35 | 1.18 |
| RB.AvgCorefChain | 0.68 | 1.19 | 1.03 | 0 | 0.34 | 1.28 | 1.1 | 0.73 |
| RB.AvgNmdEntSen | 0.67 | 0.27 | 0.42 | 0 | 0.17 | 0.72 | 2.16 | 1 |
| RB.AvgPronBl_indefinite | 0.65 | 1.4 | 1.66 | 0 | 1.98 | 1.31 | 0.13 | 0.27 |
| RB.AvgDepsBl_det | 0.65 | 1.18 | 0.96 | 0 | 0.43 | 0.97 | 0.92 | 0.91 |
| RB.AvgDepsBl_dobj | 0.65 | 1.4 | 0.87 | 0 | 0.37 | 1.26 | 0 | 1.73 |
| RB.SynDiv | 0.6 | 0.64 | 0.7 | 0 | 0.42 | 0.71 | 1.55 | 0.64 |
| RB.AvgAOEBl_InflectionPointPolynomial | 0.6 | 0.88 | 0.95 | 0 | 2.01 | 1.2 | 0 | 1.09 |
| RB.FrqRhythmId | 0.59 | 1.12 | 1.27 | 0 | 0.11 | 0.77 | 1.31 | 0.18 |
| RB.LangRhythmDiameter | 0.58 | 0.18 | 0.35 | 0 | 0.17 | 0.01 | 2.28 | 0.82 |
| RB.AvgDepsBl_expl | 0.58 | 0.64 | 0.28 | 0 | 0.35 | 0.2 | 1.79 | 0.82 |
| RB.CharEnt | 0.57 | 1.37 | 1.01 | 0 | 0.53 | 1.35 | 0.86 | 0 |
| RB.AvgNounSen | 0.55 | 0.74 | 0.91 | 0 | 0.31 | 0.46 | 1.46 | 0.36 |
| RB.AvgDepsBl_amod | 0.55 | 0.92 | 0.33 | 0 | 0.1 | 0.55 | 1.08 | 1.09 |
| RB.AvgUnqVerbBl | 0.54 | 1.56 | 1.01 | 0 | 0.48 | 1.48 | 0.16 | 0.36 |
| RB.AvgUnqPronounBl | 0.54 | 1.6 | 0.94 | 0 | 1.5 | 1.7 | 0 | 0 |
| RB.AvgPronBl_first_person | 0.53 | 1.35 | 0.7 | 0 | 0.34 | 1.23 | 0.61 | 0.36 |
| RB.AvgConnBl_sentence_linking | 0.53 | 1.45 | 1.07 | 0 | 0.41 | 1.41 | 0 | 0.64 |
| RB.LxcSoph | 0.52 | 0.32 | 0.77 | 0 | 0.46 | 0.6 | 0.1 | 2.09 |
| RB.AvgAOEBl_IndexPolynomialFitAboveThreshold.0.3. | 0.5 | 0.94 | 1.01 | 0 | 0.41 | 1.09 | 0.73 | 0.27 |
| RB.AvgRhythmUnitStreesSyll | 0.49 | 0.65 | 0.67 | 0 | 0.45 | 0.43 | 0.75 | 1 |
| RB.AvgDepsBl_mark | 0.47 | 1.36 | 0.95 | 0 | 0.08 | 1.19 | 0 | 0.64 |
| RB.AvgDepsBl_nmod | 0.47 | 1.27 | 0.79 | 0 | 0.43 | 1.1 | 0 | 0.73 |
| RB.WdDiffLemmaStem | 0.45 | 0.64 | 0.88 | 0 | 0.65 | 1.02 | 0.74 | 0.18 |
| RB.AvgDepsBl_conj | 0.45 | 0.95 | 0.52 | 0 | 0.28 | 0.64 | 0.41 | 0.91 |
| RB.AvgAOABl_Bird | 0.44 | 0.56 | 0.39 | 0 | 0.58 | 0.57 | 0.96 | 0.55 |
| RB.AvgPronBl_third_person | 0.43 | 1.36 | 1.14 | 0 | 0.33 | 1.26 | 0 | 0.09 |
| RB.AvgDepsBl_ccomp | 0.43 | 0.93 | 0.86 | 0 | 0.04 | 0.47 | 1.06 | 0 |
| RB.AggPronSen_third_person | 0.43 | 0.52 | 0.51 | 0 | 0.11 | 1.01 | 1.15 | 0.18 |
| RB.AvgDepsSen_punct | 0.43 | 0.38 | 0.62 | 0 | 0.19 | 0.94 | 0.81 | 0.64 |
| RB.AvgConnBl_simple_subordinators | 0.42 | 1.31 | 1.02 | 0 | 0.73 | 1.08 | 0 | 0.09 |
| RB.AvgConnSen_simple_subordinators | 0.41 | 0.15 | 0.52 | 0 | 0.09 | 0.52 | 1.54 | 0.18 |
| RB.AvgSenBlCoh_LDA | 0.4 | 0.74 | 0.95 | 0 | 0.2 | 1.21 | 0 | 0.73 |
| RB.AvgDepsBl_xcomp | 0.4 | 1.15 | 0.82 | 0 | 0.37 | 0.88 | 0.45 | 0 |
| RB.AvgCommaBl | 0.4 | 0.72 | 0.45 | 0 | 0.05 | 0.39 | 0.96 | 0.45 |
| RB.AvgAOASen_Shock | 0.39 | 0.4 | 0.74 | 0 | 0.45 | 0.9 | 0.79 | 0.18 |
| RB.AvgSenBlCoh_word2vec | 0.36 | 1.11 | 0.78 | 0 | 0.18 | 1.03 | 0 | 0.27 |
| RB.WdLettStdDev | 0.34 | 0.72 | 0.63 | 0 | 0.39 | 0.7 | 0.46 | 0.18 |
| RB.AvgConnBl_temporal_connectors | 0.34 | 1.03 | 0.92 | 0 | 0.02 | 0.77 | 0.1 | 0.27 |
| RB.AvgDepsBl_acl | 0.34 | 0.58 | 0.44 | 0 | 0.07 | 0.2 | 1.19 | 0 |
| RB.LangRhythmCoeff | 0.33 | 0.7 | 0.59 | 0 | 0.25 | 0.66 | 0.61 | 0 |
| RB.WdSylCnt | 0.33 | 0.38 | 0.9 | 0 | 0.27 | 0.79 | 0.26 | 0.45 |
| RB.AvgDepsBl_auxpass | 0.33 | 0.86 | 0.55 | 0 | 0.01 | 0.5 | 0.7 | 0 |
| RB.AvgConnBl_oppositions | 0.33 | 0.85 | 0.49 | 0 | 0 | 0.42 | 0.63 | 0.18 |
| RB.AvgAdverbBl | 0.32 | 1.1 | 0.72 | 0 | 0.11 | 0.83 | 0 | 0.18 |
| RB.AvgConnBl_order | 0.32 | 0.73 | 0.27 | 0 | 0.01 | 0.29 | 1 | 0 |
| RB.AvgAOABl_Bristol | 0.31 | 0.75 | 0.29 | 0 | 0.7 | 0.77 | 0.39 | 0 |
| RB.AvgDepsSen_nmod | 0.31 | 0.06 | 0.43 | 0 | 0.11 | 0.34 | 0.45 | 1 |
| RB.AvgPronounSen | 0.31 | 0.36 | 0.7 | 0 | 0.05 | 0.55 | 0 | 1 |
| RB.AvgIntraBlCoh_Path | 0.3 | 1.14 | 0.3 | 0 | 0.16 | 0.97 | 0 | 0.18 |
| RB.AvgAOABl_Kuperman | 0.3 | 0.51 | 0.51 | 0 | 0.71 | 0.59 | 0.1 | 0.45 |
| RB.AvgDepsSen_nsubj | 0.3 | 0.05 | 0.83 | 0 | 0.04 | 0.49 | 0 | 1.18 |
| RB.AvgDepsSen_aux | 0.3 | 0.17 | 0.62 | 0 | 0.21 | 0.49 | 0.68 | 0.36 |
| RB.AvgInferenceDistChain | 0.29 | 0.8 | 0.71 | 0 | 0.19 | 0.74 | 0.25 | 0 |
| RB.AvgConnBl_conditions | 0.29 | 0.9 | 0.45 | 0 | 0.15 | 0.49 | 0.44 | 0 |
| RB.AvgDepsBl_cop | 0.28 | 1.07 | 0.53 | 0 | 0.08 | 0.7 | 0 | 0.18 |
| RB.RdbltyFlesch | 0.28 | 0.49 | 0.88 | 0 | 0.38 | 0.54 | 0 | 0.45 |
| RB.AvgConnSen_temporal_connectors | 0.28 | 0.28 | 0.82 | 0 | 0.17 | 0.05 | 0.75 | 0.18 |
| RB.AvgUnqAdjectiveBl | 0.27 | 1.2 | 0.24 | 0 | 0.01 | 0.97 | 0.03 | 0 |
| RB.AvgDepsBl_advcl | 0.27 | 1.18 | 0.37 | 0 | 0.08 | 0.9 | 0.01 | 0 |
| RB.AvgDepsSen_advcl | 0.27 | 0.16 | 0.53 | 0 | 0.12 | 0.69 | 0.66 | 0.18 |
| RB.AggPronSen_indefinite | 0.25 | 0.42 | 0.84 | 0 | 0.26 | 1.17 | 0.03 | 0 |
| RB.WdDiffWdStem | 0.25 | 0.65 | 0.56 | 0 | 0.42 | 0.81 | 0.13 | 0 |
| RB.AvgDepsBl_neg | 0.24 | 0.45 | 0.08 | 0 | 0.02 | 0.12 | 0.9 | 0 |
| RB.AvgDepsBl_nummod | 0.23 | 0.45 | 0.11 | 0 | 0 | 0.12 | 0.89 | 0 |
| RB.AvgDepsBl_mwe | 0.22 | 0.29 | 0.46 | 0 | 0 | 0.06 | 0.76 | 0 |
| RB.AvgDepsSen_amod | 0.22 | 0.3 | 0.64 | 0 | 0.32 | 0.72 | 0.22 | 0 |
| RB.AvgAOASen_Bird | 0.21 | 0.32 | 0.74 | 0 | 0.27 | 0.42 | 0.25 | 0 |
| RB.AvgPrepositionSen | 0.21 | 0.07 | 0.66 | 0 | 0.05 | 0.35 | 0 | 0.73 |
| RB.AvgConnBl_contrasts | 0.21 | 1.04 | 0.19 | 0 | 0.01 | 0.64 | 0 | 0 |
| RB.AvgAOASen_Kuperman | 0.21 | 0.5 | 0.2 | 0 | 0.88 | 0.48 | 0 | 0.18 |
| RB.AvgDepsSen_xcomp | 0.21 | 0.19 | 0.71 | 0 | 0.05 | 1.01 | 0.23 | 0 |
| RB.AvgDepsBl_root | 0.2 | 0.04 | 0.29 | 0 | 0.02 | 0 | 0.99 | 0 |
| RB.AvgDepsSen_cop | 0.2 | 0.06 | 0.49 | 0 | 0.28 | 0.36 | 0.6 | 0 |
| RB.AvgConnSen_reason_and_purpose | 0.19 | 0.14 | 0.21 | 0 | 0.11 | 0.61 | 0.53 | 0 |
| RB.AvgDepsSen_conj | 0.19 | 0.14 | 0.56 | 0 | 0.02 | 0.42 | 0 | 0.55 |
| RB.AvgDepsSen_dobj | 0.19 | 0.06 | 0.6 | 0 | 0.23 | 0.38 | 0 | 0.55 |
| RB.AvgDepsSen_dep | 0.19 | 0.49 | 0.57 | 0 | 0.29 | 0.65 | 0 | 0 |
| RB.AvgAdverbSen | 0.19 | 0 | 0.72 | 0 | 0.05 | 0.87 | 0 | 0.36 |
| RB.AvgSenLen | 0.18 | 0.06 | 0.76 | 0 | 0.12 | 0.29 | 0 | 0.45 |
| RB.AvgPronBl_second_person | 0.18 | 0.7 | 0.62 | 0 | 0.01 | 0.3 | 0 | 0 |
| RB.AvgConnBl_disjunctions | 0.18 | 0.73 | 0.25 | 0 | 0.01 | 0.35 | 0.16 | 0 |
| RB.AvgConnBl_reason_and_purpose | 0.18 | 0.64 | 0.26 | 0 | 0.03 | 0.26 | 0.28 | 0 |
| RB.AggPronSen_second_person | 0.18 | 0.32 | 0.47 | 0 | 0.01 | 0.08 | 0.53 | 0 |
| RB.AvgConnBl_semi_coordinators | 0.16 | 0.35 | 0.31 | 0 | 0.01 | 0.09 | 0.43 | 0 |
| RB.AvgPronBl_interrogative | 0.16 | 0.7 | 0.35 | 0 | 0.01 | 0.28 | 0.07 | 0 |
| RB.AvgAOASen_Bristol | 0.15 | 0.41 | 0.39 | 0 | 0.14 | 0.58 | 0 | 0 |
| RB.AvgDepsSen_ccomp | 0.15 | 0.18 | 0.59 | 0 | 0.11 | 0.47 | 0 | 0.18 |
| RB.AvgDepsBl_iobj | 0.15 | 0.62 | 0.33 | 0 | 0.01 | 0.29 | 0 | 0.09 |
| RB.AvgDepsSen_det | 0.15 | 0.21 | 0.32 | 0 | 0.24 | 0.07 | 0.12 | 0.36 |
| RB.AvgConnSen_addition | 0.13 | 0.11 | 0.27 | 0 | 0.75 | 0.46 | 0 | 0 |
| RB.AvgDepsSen_acl | 0.13 | 0.38 | 0.58 | 0 | 0.11 | 0.08 | 0 | 0.09 |
| RB.AvgDepsSen_mark | 0.12 | 0.09 | 0.35 | 0 | 0.04 | 0.36 | 0 | 0.27 |
| RB.AvgConnSen_oppositions | 0.12 | 0.25 | 0.65 | 0 | 0.08 | 0.05 | 0.13 | 0 |
| RB.AvgDepsBl_dep | 0.11 | 0.58 | 0.04 | 0 | 0.06 | 0.19 | 0.04 | 0 |
| RB.AvgConnSen_semi_coordinators | 0.11 | 0.22 | 0.65 | 0 | 0.23 | 0.04 | 0 | 0 |
| RB.AvgConnBl_complex_subordinators | 0.11 | 0.39 | 0.19 | 0 | 0 | 0.12 | 0.19 | 0 |
| RB.AvgAOASen_Cortese | 0.11 | 0.12 | 0.25 | 0 | 0.35 | 0.64 | 0 | 0 |
| RB.AvgAdjectiveSen | 0.1 | 0.09 | 0.4 | 0 | 0.05 | 0.56 | 0 | 0 |
| RB.AvgDepsSen_iobj | 0.07 | 0.16 | 0.45 | 0 | 0.02 | 0.02 | 0 | 0 |
| RB.AggPronSen_interrogative | 0.07 | 0.11 | 0.4 | 0 | 0.21 | 0.01 | 0 | 0 |
| RB.AvgConnSen_order | 0.07 | 0.02 | 0.48 | 0 | 0.13 | 0 | 0 | 0.09 |
| RB.SenAsson | 0.07 | 0.24 | 0.41 | 0 | 0 | 0.02 | 0 | 0 |
| RB.AvgConnSen_conditions | 0.06 | 0.07 | 0.49 | 0 | 0.08 | 0 | 0 | 0 |
| RB.AvgDepsBl_csubj | 0.06 | 0.02 | 0.31 | 0 | 0.06 | 0 | 0.17 | 0 |
| RB.AvgDepsSen_neg | 0.06 | 0.17 | 0.4 | 0 | 0.03 | 0.03 | 0 | 0 |
| RB.AvgDepsBl_parataxis | 0.05 | 0.24 | 0.12 | 0 | 0 | 0.04 | 0 | 0 |
| RB.AvgDepsBl_appos | 0.04 | 0.2 | 0.08 | 0 | 0 | 0.04 | 0 | 0 |
| RB.AvgDepsSen_nummod | 0.04 | 0.04 | 0.35 | 0 | 0.02 | 0 | 0 | 0 |
| RB.AggPronSen_first_person | 0.04 | 0.02 | 0.2 | 0 | 0.14 | 0.1 | 0 | 0 |
| RB.AvgConnSen_disjunctions | 0.03 | 0.11 | 0.13 | 0 | 0.04 | 0.01 | 0 | 0 |
| RB.SenAllit | 0.03 | 0.03 | 0.3 | 0 | 0 | 0 | 0 | 0 |
| RB.AvgDepsSen_mwe | 0.01 | 0.07 | 0 | 0 | 0 | 0.01 | 0 | 0 |
ReaderBench Model 3exp
This model was trained on 15 min expository writing samples.
Algorithm Weightings in Ensemble
Abbreviations: * overall = ensemble model * pls = partial least squares regression * gbm = stochastic gradient boosted trees * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | rf | mars | gbm | svm | enet | cube |
|---|---|---|---|---|---|---|
| -0.0156 | 0.0826 | 0.3112 | 0.0319 | 0.1360 | 0.3306 | 0.1259 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | rf | mars | gbm | svm | enet | cube |
|---|---|---|---|---|---|---|---|
| Content.words | 20.83 | 5.13 | 35.84 | 48.17 | 3.43 | 17.49 | 14.66 |
| RB.AvgWdLen | 4.5 | 1.29 | 10.64 | 1.06 | 0.42 | 1.6 | 4.32 |
| RB.AvgDepsBl_compound | 4.11 | 0.71 | 8.06 | 0.36 | 0.06 | 3.36 | 3.92 |
| RB.AvgConnBl_order | 3.63 | 0.6 | 6.17 | 0.15 | 0.21 | 4.43 | 1.81 |
| RB.SenStdDevWd | 3.56 | 1.02 | 5.2 | 0.29 | 1.18 | 4.38 | 2.41 |
| RB.LangRhythmId | 3.46 | 0.96 | 10.64 | 0.29 | 0.26 | 0 | 0.7 |
| RB.TCorefChainDoc | 3.34 | 1.93 | 0 | 4.43 | 2.48 | 7.01 | 3.51 |
| RB.WdEnt | 3.21 | 2.23 | 0 | 1.79 | 2.35 | 6.08 | 5.52 |
| RB.AggPronSen_first_person | 2.93 | 0.99 | 8.76 | 0.77 | 0.12 | 0 | 1.1 |
| Sentences | 2.9 | 1.37 | 0 | 2.38 | 1.47 | 6.99 | 2.01 |
| RB.AvgSenAdjCoh_Path | 2.68 | 1.05 | 0 | 1.28 | 1.09 | 5.93 | 3.92 |
| RB.CAF | 2.51 | 1.33 | 0 | 1.24 | 1.75 | 5.87 | 1.81 |
| RB.AvgPronBl_third_person | 2.49 | 0.76 | 7.52 | 0.04 | 0.79 | 0 | 0.2 |
| RB.AvgBlScore | 2.27 | 2.1 | 4.33 | 1.67 | 2.38 | 0 | 3.31 |
| RB.AvgPronBl_second_person | 2.03 | 1.17 | 0 | 0.64 | 0.95 | 4.73 | 2.01 |
| RB.LangRhythmDiameter | 1.92 | 0.73 | 2.84 | 0.28 | 0.15 | 2.24 | 1.91 |
| RB.TActCorefChainWd | 1.4 | 0.76 | 0 | 0.47 | 1.04 | 2.97 | 1.81 |
| RB.TCorefChainBigSpan | 1.31 | 0.44 | 0 | 1.17 | 1.63 | 2.75 | 1 |
| RB.AvgUnqAdjectiveBl | 1.06 | 1.01 | 0 | 0.23 | 1.7 | 1.95 | 0.9 |
| RB.WdDiffWdStem | 0.99 | 1.06 | 0 | 1.8 | 0.52 | 2.17 | 0.6 |
| RB.AvgDepsSen_nmod | 0.94 | 0.95 | 0 | 0.52 | 1.14 | 1.67 | 1.2 |
| RB.AvgDepsBl_expl | 0.89 | 0.79 | 0 | 0.42 | 0.42 | 1.86 | 1.2 |
| RB.RdbltyDaleChall | 0.86 | 1.25 | 0 | 0.91 | 0.78 | 1.01 | 2.41 |
| RB.AvgAOEBl_InflectionPointPolynomial | 0.77 | 0.72 | 0 | 0.27 | 0.7 | 1.85 | 0.1 |
| RB.AvgConnBl_temporal_connectors | 0.76 | 0.91 | 0 | 0.04 | 0.5 | 1.37 | 1.41 |
| RB.AvgPronBl_indefinite | 0.75 | 2.03 | 0 | 5.56 | 1.56 | 0 | 1.61 |
| RB.SynDiv | 0.71 | 0.61 | 0 | 0.28 | 1.04 | 1.39 | 0.5 |
| RB.LxcDiv | 0.69 | 1.51 | 0 | 1.57 | 2.14 | 0 | 1.91 |
| RB.AvgAOASen_Bristol | 0.66 | 0.56 | 0 | 0.14 | 0.31 | 1.56 | 0.5 |
| RB.AvgDepsBl_root | 0.65 | 0.09 | 0 | 0.04 | 0.04 | 1.95 | 0 |
| RB.AvgDepsBl_nsubj | 0.62 | 1.9 | 0 | 0.88 | 2.19 | 0 | 1.2 |
| RB.AvgPronounBl | 0.59 | 1.54 | 0 | 0.25 | 1.94 | 0 | 1.61 |
| RB.AvgPrepositionBl | 0.59 | 1.37 | 0 | 1.41 | 2.07 | 0 | 1.31 |
| RB.AvgUnqNoundBl | 0.49 | 0.83 | 0 | 0.41 | 1.02 | 0 | 2.21 |
| RB.AvgDepsBl_parataxis | 0.47 | 0.53 | 0 | 0.01 | 0.15 | 1.26 | 0 |
| RB.LangRhythmCoeff | 0.44 | 0.58 | 0 | 0.33 | 0.4 | 0.93 | 0.2 |
| RB.AvgUnqPrepositionBl | 0.43 | 0.94 | 0 | 0.2 | 2.05 | 0 | 0.6 |
| RB.AvgAOASen_Bird | 0.43 | 0.63 | 0 | 0.4 | 0.79 | 0.63 | 0.5 |
| RB.WdSylCnt | 0.42 | 0.96 | 0 | 0.54 | 0.18 | 0.5 | 1.1 |
| RB.AvgDepsBl_nmod | 0.42 | 1.01 | 0 | 0.52 | 1.59 | 0 | 0.9 |
| RB.AvgChainSpan | 0.41 | 1.04 | 0 | 0.4 | 1.58 | 0 | 0.8 |
| RB.AvgDepsBl_nummod | 0.41 | 0.7 | 0 | 0.01 | 0.21 | 1 | 0 |
| RB.AvgDepsSen_expl | 0.4 | 0.41 | 0 | 0.23 | 0.06 | 1.1 | 0 |
| RB.AvgPronBl_first_person | 0.39 | 0.71 | 0 | 0.51 | 0.5 | 0.25 | 1.41 |
| RB.AvgUnqVerbBl | 0.38 | 0.91 | 0 | 0.06 | 1.71 | 0 | 0.6 |
| RB.AvgDepsBl_aux | 0.37 | 0.59 | 0 | 0.19 | 0.93 | 0.4 | 0.5 |
| RB.AvgAdverbBl | 0.33 | 0.6 | 0 | 0.11 | 1.31 | 0 | 0.8 |
| RB.AvgDepsBl_punct | 0.33 | 1.26 | 0 | 0.3 | 1.17 | 0 | 0.5 |
| RB.AvgNounSen | 0.33 | 0.99 | 0 | 0.05 | 0.22 | 0 | 1.81 |
| RB.LxcSoph | 0.32 | 0.79 | 0 | 0.3 | 0.75 | 0 | 1.2 |
| RB.CharEnt | 0.31 | 0.49 | 0 | 1.05 | 1.09 | 0.13 | 0.4 |
| RB.AvgDepsSen_cop | 0.31 | 0.86 | 0 | 0.55 | 0.55 | 0 | 1.2 |
| RB.AvgDepsBl_mark | 0.31 | 1.04 | 0 | 0.56 | 1.59 | 0 | 0 |
| RB.AvgSenBlCoh_LDA | 0.3 | 0.82 | 0 | 0.16 | 1.15 | 0 | 0.6 |
| RB.RdbltyFlesch | 0.29 | 0.47 | 0 | 0.19 | 0.17 | 0 | 1.81 |
| RB.AvgCorefChain | 0.28 | 0.76 | 0 | 0.2 | 1.05 | 0 | 0.6 |
| RB.AvgDepsBl_dobj | 0.28 | 0.92 | 0 | 0.09 | 1.36 | 0 | 0.2 |
| RB.AvgDepsBl_cop | 0.27 | 0.59 | 0 | 0.07 | 0.97 | 0 | 0.7 |
| RB.AvgDepsBl_det | 0.27 | 0.92 | 0 | 0.09 | 1.36 | 0 | 0.1 |
| RB.AvgDepsSen_mark | 0.27 | 0.68 | 0 | 0.19 | 1.12 | 0 | 0.5 |
| RB.AvgDepsBl_amod | 0.26 | 0.58 | 0 | 0.27 | 1.23 | 0 | 0.3 |
| RB.AvgDepsBl_mwe | 0.25 | 0.8 | 0 | 0.09 | 0.61 | 0.3 | 0 |
| RB.AvgUnqAdverbBl | 0.25 | 0.6 | 0 | 0.03 | 1.39 | 0 | 0.1 |
| RB.AvgPrepositionSen | 0.24 | 0.44 | 0 | 0.16 | 0.91 | 0 | 0.6 |
| RB.AvgConnBl_simple_subordinators | 0.23 | 0.76 | 0 | 0.05 | 1.22 | 0 | 0 |
| RB.AvgAOASen_Kuperman | 0.23 | 0.53 | 0 | 0.51 | 0.39 | 0.2 | 0.4 |
| RB.AvgDepsSen_compound | 0.23 | 1.22 | 0 | 0.33 | 0.33 | 0 | 0.6 |
| RB.AvgDepsBl_ccomp | 0.22 | 0.51 | 0 | 0.05 | 0.54 | 0.2 | 0.3 |
| RB.AvgUnqPronounBl | 0.22 | 0.46 | 0 | 0 | 1.33 | 0 | 0 |
| RB.FrqRhythmId | 0.22 | 0.94 | 0 | 0.3 | 0.68 | 0.06 | 0.2 |
| RB.AggPronSen_indefinite | 0.22 | 0.76 | 0 | 0.37 | 0.93 | 0 | 0.2 |
| RB.AvgDepsSen_dobj | 0.21 | 0.98 | 0 | 0.1 | 0.49 | 0 | 0.5 |
| RB.AggPronSen_second_person | 0.2 | 0.81 | 0 | 0.23 | 0.64 | 0 | 0.3 |
| RB.AvgAOADoc_Shock | 0.2 | 0.98 | 0 | 0.42 | 0.82 | 0 | 0 |
| RB.AvgConnSen_semi_coordinators | 0.19 | 0.59 | 0 | 0.29 | 0 | 0.38 | 0.1 |
| RB.AvgConnBl_addition | 0.18 | 0.7 | 0 | 0.23 | 0.65 | 0 | 0.2 |
| RB.AvgRhythmUnitStreesSyll | 0.18 | 0.89 | 0 | 0.17 | 0.47 | 0 | 0.3 |
| RB.AvgDepsSen_ccomp | 0.18 | 0.31 | 0 | 0.22 | 0.94 | 0 | 0.2 |
| RB.AvgAdverbSen | 0.17 | 0.38 | 0 | 0.06 | 0.99 | 0 | 0 |
| RB.AvgCommaSen | 0.17 | 0.62 | 0 | 0.25 | 0.8 | 0 | 0 |
| RB.AvgAOEDoc_IndexAboveThreshold.0.3. | 0.17 | 0.72 | 0 | 0.12 | 0.36 | 0 | 0.5 |
| RB.AvgConnBl_contrasts | 0.17 | 0.46 | 0 | 0.08 | 0.82 | 0 | 0.2 |
| RB.AvgConnSen_simple_subordinators | 0.16 | 0.44 | 0 | 0.13 | 0.88 | 0 | 0 |
| RB.AvgConnBl_reason_and_purpose | 0.16 | 0.73 | 0 | 0.14 | 0.62 | 0 | 0.1 |
| RB.AvgAOADoc_Bird | 0.16 | 0.79 | 0 | 0.14 | 0.68 | 0 | 0 |
| RB.AvgDepsSen_amod | 0.16 | 0.29 | 0 | 0.25 | 0.5 | 0 | 0.5 |
| RB.AvgConnBl_oppositions | 0.16 | 0.65 | 0 | 0.05 | 0.6 | 0.02 | 0.2 |
| RB.AvgAOABl_Kuperman | 0.15 | 0.11 | 0 | 0.18 | 0.45 | 0 | 0.6 |
| RB.AvgDepsSen_xcomp | 0.15 | 0.63 | 0 | 0.06 | 0.73 | 0 | 0 |
| RB.AvgPronounSen | 0.14 | 0.62 | 0 | 0.03 | 0.26 | 0 | 0.4 |
| RB.AvgDepsBl_advcl | 0.14 | 0.21 | 0 | 0.02 | 0.89 | 0 | 0 |
| RB.AvgInferenceDistChain | 0.14 | 0.56 | 0 | 0.2 | 0.45 | 0 | 0.2 |
| RB.AvgNounNmdEntBl | 0.14 | 0.49 | 0 | 0.87 | 0.55 | 0 | 0 |
| RB.AggPronSen_third_person | 0.14 | 0.65 | 0 | 0.14 | 0.64 | 0 | 0 |
| RB.WdLettStdDev | 0.14 | 0.65 | 0 | 0.18 | 0.63 | 0 | 0 |
| RB.AvgConnSen_addition | 0.13 | 0.47 | 0 | 0.23 | 0.63 | 0 | 0 |
| RB.AvgNmdEntSen | 0.13 | 0.18 | 0 | 0.36 | 0.81 | 0 | 0 |
| RB.WdDiffLemmaStem | 0.12 | 0.71 | 0 | 0.26 | 0.29 | 0 | 0.1 |
| RB.AvgDepsSen_aux | 0.12 | 0.4 | 0 | 0.03 | 0.64 | 0 | 0 |
| RB.AvgCommaBl | 0.12 | 0.66 | 0 | 0.04 | 0.4 | 0 | 0.1 |
| RB.AvgAOASen_Shock | 0.12 | 0.28 | 0 | 0.05 | 0.73 | 0 | 0 |
| RB.AvgDepsBl_acl | 0.12 | 0.47 | 0 | 0.13 | 0.6 | 0 | 0 |
| RB.AvgAOABl_Cortese | 0.12 | 0.28 | 0 | 0.1 | 0.64 | 0 | 0.1 |
| RB.AvgDepsSen_advcl | 0.12 | 0.46 | 0 | 0.25 | 0.59 | 0 | 0 |
| RB.AvgDepsBl_xcomp | 0.12 | 0.23 | 0 | 0.09 | 0.78 | 0 | 0 |
| RB.AvgConnSen_temporal_connectors | 0.11 | 0.73 | 0 | 0.06 | 0.09 | 0.06 | 0.1 |
| RB.AvgAOESen_InflectionPointPolynomial | 0.11 | 0.28 | 0 | 0.11 | 0.52 | 0 | 0.1 |
| RB.AvgDepsSen_dep | 0.11 | 0.49 | 0 | 0.17 | 0.38 | 0 | 0.1 |
| RB.AvgAOASen_Cortese | 0.11 | 0.22 | 0 | 0.17 | 0.66 | 0 | 0 |
| RB.AvgDepsSen_det | 0.11 | 0.14 | 0 | 0.12 | 0.54 | 0 | 0.2 |
| RB.AvgConnSen_reason_and_purpose | 0.11 | 0.39 | 0 | 0.12 | 0.58 | 0 | 0 |
| RB.AvgAOABl_Bristol | 0.1 | 0.45 | 0 | 0.15 | 0.37 | 0 | 0.1 |
| RB.AvgDepsBl_iobj | 0.09 | 0.74 | 0 | 0.21 | 0.17 | 0 | 0 |
| RB.AvgDepsSen_mwe | 0.09 | 0.64 | 0 | 0.44 | 0.21 | 0 | 0 |
| RB.AvgConnSen_order | 0.08 | 0.69 | 0 | 0.67 | 0.01 | 0 | 0 |
| RB.AvgConnSen_oppositions | 0.08 | 0.63 | 0 | 0.11 | 0.09 | 0 | 0.1 |
| RB.AvgConnBl_disjunctions | 0.08 | 0.5 | 0 | 0 | 0.32 | 0 | 0 |
| RB.AvgConnSen_contrasts | 0.07 | 0.6 | 0 | 0.11 | 0.11 | 0 | 0 |
| RB.AvgDepsBl_auxpass | 0.07 | 0.56 | 0 | 0.01 | 0.17 | 0 | 0 |
| RB.AvgDepsSen_neg | 0.07 | 0.47 | 0 | 0.31 | 0 | 0 | 0.2 |
| RB.AvgConnBl_conditions | 0.07 | 0.49 | 0 | 0.11 | 0.23 | 0 | 0 |
| RB.AvgDepsBl_neg | 0.06 | 0.19 | 0 | 0.03 | 0.23 | 0 | 0.1 |
| RB.AvgPronBl_interrogative | 0.06 | 0.54 | 0 | 0.04 | 0.14 | 0 | 0 |
| RB.SenAsson | 0.05 | 0.25 | 0 | 0 | 0.23 | 0 | 0 |
| RB.AvgConnSen_disjunctions | 0.05 | 0.55 | 0 | 0.03 | 0.05 | 0 | 0 |
| RB.AvgConnBl_semi_coordinators | 0.04 | 0.16 | 0 | 0.1 | 0.16 | 0 | 0 |
| RB.AvgDepsSen_nummod | 0.04 | 0.46 | 0 | 0.03 | 0 | 0 | 0 |
| RB.AvgDepsSen_acl | 0.04 | 0.46 | 0 | 0.01 | 0.01 | 0 | 0 |
| RB.AvgDepsBl_csubj | 0.04 | 0.4 | 0 | 0.01 | 0.05 | 0 | 0 |
| RB.AvgDepsBl_nsubjpass | 0.04 | 0.25 | 0 | 0 | 0.16 | 0 | 0 |
| RB.AvgDepsBl_appos | 0.04 | 0.51 | 0 | 0 | 0.02 | 0 | 0 |
| RB.AvgDepsBl_dep | 0.02 | 0 | 0 | 0.08 | 0.15 | 0 | 0 |
| RB.SenAllit | 0.02 | 0.3 | 0 | 0 | 0 | 0 | 0 |
ReaderBench Model 3per
This modelwas trained on 15 min persuasive writing samples.
Algorithm Weightings in Ensemble
Abbreviations: * overall = ensemble model * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | mars | gbm | svm | enet | cube |
|---|---|---|---|---|---|---|
| -0.0141 | 0.0326 | 0.2043 | 0.2331 | 0.1507 | 0.3202 | 0.0801 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | mars | gbm | svm | enet | cube |
|---|---|---|---|---|---|---|---|
| RB.WdEnt | 9.44 | 1.97 | 0 | 16.45 | 2.58 | 14.62 | 8.38 |
| RB.AvgPrepositionBl | 8.44 | 1.96 | 20.57 | 11.55 | 2.48 | 2.8 | 4.83 |
| Sentences | 6.71 | 1.67 | 19.13 | 4.09 | 1.41 | 4.14 | 5.01 |
| RB.AvgBlScore | 5.39 | 2 | 0 | 15.75 | 2.73 | 2.76 | 5.92 |
| RB.CAF | 4.59 | 1.59 | 19.13 | 1.27 | 1.43 | 0 | 2.73 |
| RB.AvgSenScore | 3.72 | 0.51 | 8.2 | 0.15 | 0.47 | 5.74 | 2 |
| RB.TCorefChainDoc | 3.36 | 1.97 | 0 | 5.74 | 2.16 | 4.69 | 2.46 |
| RB.AvgWdLen | 2.49 | 1.32 | 0 | 4.98 | 1.13 | 2.84 | 3.28 |
| RB.AvgAOADoc_Shock | 2.39 | 1.4 | 8.2 | 1.65 | 1.21 | 0.2 | 1.18 |
| RB.AvgPronBl_indefinite | 2.34 | 1.76 | 0 | 3.91 | 2.01 | 2.57 | 3.73 |
| RB.RdbltyDaleChall | 2.32 | 0.79 | 0 | 1.32 | 0.68 | 5.11 | 3.73 |
| RB.AvgDepsBl_compound | 2.28 | 0.23 | 7.3 | 0.29 | 0.02 | 1.88 | 2 |
| RB.AvgUnqNoundBl | 2.11 | 0.75 | 1.47 | 0.3 | 1.41 | 4.17 | 2.64 |
| RB.AvgConnBl_simple_subordinators | 1.75 | 1.76 | 0 | 3.08 | 1.98 | 2.11 | 0.46 |
| RB.AvgAOESen_InflectionPointPolynomial | 1.61 | 0.53 | 5.45 | 0.22 | 0.77 | 0.99 | 0.36 |
| RB.AvgPronBl_interrogative | 1.23 | 0.61 | 0 | 0.53 | 0.13 | 2.92 | 2 |
| RB.AvgDepsBl_nsubj | 1.12 | 1.87 | 0 | 1.87 | 2.47 | 0 | 3.37 |
| RB.AvgDepsBl_mark | 1.1 | 1.63 | 0 | 0.89 | 1.7 | 1.41 | 1.91 |
| RB.AvgNmdEntSen | 1.07 | 0.07 | 0 | 0.25 | 0.41 | 2.79 | 0.91 |
| RB.AvgDepsBl_amod | 1.06 | 0.82 | 0 | 0.33 | 0.41 | 2.72 | 0.64 |
| RB.AvgCorefChain | 1.04 | 0.95 | 0 | 0.08 | 1.1 | 2.38 | 1 |
| RB.AvgDepsSen_advmod | 1.01 | 0.09 | 0 | 0.22 | 0.22 | 2.68 | 1 |
| RB.AvgPronBl_first_person | 0.96 | 0.7 | 2.97 | 0.06 | 0.27 | 0.75 | 0.64 |
| RB.LangRhythmCoeff | 0.95 | 0.77 | 0 | 1.43 | 0.81 | 1.35 | 0.73 |
| RB.AvgAOABl_Bird | 0.93 | 0.47 | 3.59 | 0.46 | 0.63 | 0 | 0 |
| RB.AvgDepsSen_aux | 0.92 | 0 | 4 | 0.21 | 0.18 | 0 | 0.55 |
| RB.AvgSenAdjCoh_Path | 0.87 | 1.19 | 0 | 2.13 | 1.26 | 0.33 | 0.73 |
| RB.AvgDepsBl_det | 0.83 | 1.51 | 0 | 0.54 | 1.48 | 1.21 | 0.73 |
| RB.AvgConnSen_oppositions | 0.8 | 0.24 | 0 | 0.41 | 0.01 | 2.1 | 0.46 |
| RB.AvgAOASen_Shock | 0.8 | 0.62 | 0 | 0.13 | 1.02 | 1.67 | 0.91 |
| RB.LxcDiv | 0.8 | 1.45 | 0 | 1.85 | 1.4 | 0 | 1.55 |
| RB.AvgUnqPronounBl | 0.77 | 1.68 | 0 | 0.64 | 1.73 | 0.63 | 1.46 |
| RB.AvgAOADoc_Cortese | 0.69 | 0.02 | 0 | 0.3 | 0.59 | 1.5 | 0.73 |
| RB.AvgUnqAdjectiveBl | 0.69 | 1.13 | 0 | 0.03 | 0.81 | 1.58 | 0.36 |
| RB.AvgDepsBl_nsubjpass | 0.69 | 0.48 | 0 | 0.02 | 0.16 | 2.05 | 0.09 |
| RB.AvgAOASen_Bird | 0.64 | 0.52 | 0 | 0.31 | 0.39 | 1.46 | 0.46 |
| RB.AvgDepsBl_cop | 0.63 | 1.09 | 0 | 0.1 | 0.8 | 1.3 | 0.55 |
| RB.TCorefChainBigSpan | 0.6 | 1.53 | 0 | 0.34 | 1.38 | 0.62 | 1 |
| RB.AvgChainSpan | 0.59 | 1.42 | 0 | 1 | 1.73 | 0 | 0.73 |
| RB.AvgDepsBl_aux | 0.59 | 1.42 | 0 | 0.59 | 1.36 | 0.5 | 0.64 |
| RB.AvgUnqPrepositionBl | 0.58 | 1.83 | 0 | 0.66 | 2.15 | 0 | 0.64 |
| RB.AggPronSen_second_person | 0.54 | 0.32 | 0 | 0.04 | 0.55 | 1.28 | 0.46 |
| RB.SynDiv | 0.51 | 1.15 | 0 | 0.48 | 1.14 | 0.52 | 0.46 |
| RB.CharEnt | 0.49 | 1.19 | 0 | 1.21 | 1.21 | 0 | 0 |
| RB.AvgAOASen_Bristol | 0.48 | 0.35 | 0 | 0.13 | 0.36 | 1.1 | 0.46 |
| RB.AvgDepsBl_punct | 0.47 | 1.51 | 0 | 0.72 | 1.39 | 0 | 0.64 |
| RB.AvgDepsBl_nmod | 0.46 | 1.6 | 0 | 0.48 | 1.72 | 0 | 0.55 |
| RB.AvgUnqVerbBl | 0.43 | 1.51 | 0 | 0.41 | 1.47 | 0 | 0.91 |
| RB.WdDiffLemmaStem | 0.42 | 0.86 | 0 | 0.43 | 0.9 | 0.48 | 0.18 |
| RB.AvgDepsSen_mark | 0.42 | 0.28 | 0 | 0.1 | 0.29 | 0.68 | 1.73 |
| RB.WdDiffWdStem | 0.42 | 0.67 | 0 | 0.31 | 0.66 | 0.71 | 0.18 |
| RB.AvgPronounBl | 0.41 | 1.67 | 0 | 0.06 | 1.67 | 0 | 1.18 |
| RB.AvgAOASen_Cortese | 0.41 | 0.08 | 0 | 0.15 | 0.3 | 0.88 | 0.73 |
| RB.AvgConnBl_temporal_connectors | 0.41 | 0.71 | 0 | 0.02 | 0.38 | 1.05 | 0 |
| RB.AvgRhythmUnitStreesSyll | 0.38 | 0.09 | 0 | 0.12 | 0.17 | 0.87 | 0.73 |
| RB.LxcSoph | 0.37 | 0.75 | 0 | 0.68 | 0.65 | 0 | 1.18 |
| RB.AvgDepsBl_ccomp | 0.34 | 1.38 | 0 | 0.08 | 1.26 | 0.12 | 0.64 |
| RB.AvgDepsSen_neg | 0.34 | 0.23 | 0 | 0.09 | 0.52 | 0.74 | 0 |
| RB.AvgPronBl_third_person | 0.34 | 1.34 | 0 | 0.39 | 1.16 | 0 | 0.46 |
| RB.AvgDepsBl_root | 0.33 | 0.09 | 0 | 0.06 | 0 | 1 | 0 |
| RB.TActCorefChainWd | 0.33 | 0.36 | 0 | 0.36 | 0.81 | 0.14 | 0.91 |
| RB.WdSylCnt | 0.3 | 0.76 | 0 | 0.54 | 0.7 | 0 | 0.64 |
| RB.AvgUnqAdverbBl | 0.29 | 1.34 | 0 | 0.09 | 1.2 | 0 | 0.64 |
| RB.AvgDepsSen_punct | 0.27 | 0.44 | 0 | 0.09 | 0.16 | 0.68 | 0 |
| RB.AvgNmdEntBl | 0.26 | 1.25 | 0 | 0.11 | 1.05 | 0 | 0.55 |
| RB.AvgConnBl_addition | 0.25 | 1.13 | 0 | 0.16 | 0.9 | 0 | 0.55 |
| RB.AvgDepsSen_compound | 0.25 | 0.5 | 0 | 0.19 | 0.56 | 0 | 1.37 |
| RB.AggPronSen_indefinite | 0.25 | 0.42 | 0 | 0.24 | 0.9 | 0 | 0.64 |
| RB.AvgDepsBl_dobj | 0.25 | 1.37 | 0 | 0.02 | 1.15 | 0 | 0.46 |
| RB.AvgConnBl_order | 0.24 | 0.56 | 0 | 0.03 | 0.21 | 0.59 | 0 |
| RB.AvgAOADoc_Bristol | 0.24 | 0.8 | 0 | 0.28 | 0.89 | 0 | 0.27 |
| RB.SenStdDevWd | 0.24 | 0.98 | 0 | 0.18 | 1.06 | 0 | 0.18 |
| RB.FrqRhythmId | 0.23 | 1.1 | 0 | 0.02 | 0.72 | 0.21 | 0.18 |
| RB.AvgDepsBl_advmod | 0.23 | 1.21 | 0 | 0.12 | 0.96 | 0 | 0.27 |
| RB.AvgDepsBl_advcl | 0.23 | 1.36 | 0 | 0.01 | 1.26 | 0 | 0 |
| RB.AvgAdverbBl | 0.23 | 1.25 | 0 | 0.1 | 1.01 | 0 | 0.27 |
| RB.AvgConnBl_logical_connectors | 0.22 | 1.14 | 0 | 0.2 | 0.89 | 0 | 0.09 |
| RB.AvgConnBl_semi_coordinators | 0.21 | 0.2 | 0 | 0.02 | 0.03 | 0.56 | 0.18 |
| RB.AvgPronounSen | 0.21 | 0.33 | 0 | 0.15 | 0.72 | 0 | 0.73 |
| RB.AvgUnqNmdEntBl | 0.21 | 1 | 0 | 0.18 | 0.65 | 0 | 0.55 |
| RB.AvgConnSen_simple_subordinators | 0.2 | 0.46 | 0 | 0.17 | 0.74 | 0 | 0.46 |
| RB.AvgSenBlCoh_LDA | 0.2 | 0.59 | 0 | 0.06 | 0.91 | 0.01 | 0.36 |
| RB.AvgConnBl_reason_and_purpose | 0.2 | 1.2 | 0 | 0.09 | 0.96 | 0 | 0 |
| RB.AvgDepsSen_amod | 0.2 | 0.18 | 0 | 0.18 | 0.62 | 0 | 0.82 |
| RB.AvgInferenceDistChain | 0.19 | 0.33 | 0 | 0.23 | 0.81 | 0 | 0.09 |
| RB.AvgAOESen_IndexPolynomialFitAboveThreshold.0.3. | 0.19 | 0.68 | 0 | 0.32 | 0.65 | 0 | 0 |
| RB.AvgSenBlCoh_LSA | 0.19 | 0.97 | 0 | 0.09 | 0.86 | 0 | 0.18 |
| RB.AvgAOEDoc_InverseAverage | 0.18 | 0.62 | 0 | 0.17 | 0.82 | 0 | 0 |
| RB.AvgAOEBl_IndexAboveThreshold.0.3. | 0.18 | 0.59 | 0 | 0.17 | 0.71 | 0.06 | 0 |
| RB.SenAllit | 0.18 | 0.53 | 0 | 0.01 | 0.19 | 0.43 | 0 |
| RB.AvgDepsSen_dep | 0.18 | 0.19 | 0 | 0.2 | 0.42 | 0 | 0.91 |
| RB.AvgDepsBl_nummod | 0.17 | 0.66 | 0 | 0.11 | 0.32 | 0.24 | 0 |
| RB.AvgDepsSen_det | 0.16 | 0.21 | 0 | 0.08 | 0.89 | 0 | 0 |
| RB.AvgDepsBl_conj | 0.16 | 1.02 | 0 | 0.11 | 0.63 | 0 | 0.09 |
| RB.AvgDepsSen_ccomp | 0.16 | 0.3 | 0 | 0.15 | 0.55 | 0 | 0.46 |
| RB.AvgConnSen_addition | 0.15 | 0.01 | 0 | 0.11 | 0.52 | 0 | 0.55 |
| RB.AvgDepsSen_acl | 0.15 | 0.15 | 0 | 0.13 | 0.01 | 0.36 | 0 |
| RB.AvgDepsBl_xcomp | 0.14 | 0.89 | 0 | 0.04 | 0.56 | 0 | 0.18 |
| RB.AvgPronBl_second_person | 0.14 | 0.91 | 0 | 0.05 | 0.55 | 0 | 0.18 |
| RB.AvgAOABl_Kuperman | 0.14 | 0.08 | 0 | 0.23 | 0.45 | 0 | 0.18 |
| RB.AvgNounSen | 0.14 | 0.2 | 0 | 0.03 | 0.22 | 0 | 1.18 |
| RB.AvgConnBl_contrasts | 0.14 | 1.03 | 0 | 0.05 | 0.67 | 0 | 0 |
| RB.WdLettStdDev | 0.13 | 0.6 | 0 | 0.19 | 0.39 | 0 | 0.09 |
| RB.AvgDepsBl_neg | 0.13 | 0.3 | 0 | 0.03 | 0.05 | 0.33 | 0 |
| RB.AvgDepsSen_xcomp | 0.13 | 0.06 | 0 | 0.05 | 0.58 | 0 | 0.36 |
| RB.AvgDepsSen_advcl | 0.13 | 0.14 | 0 | 0.13 | 0.66 | 0 | 0 |
| RB.AvgConnBl_oppositions | 0.13 | 0.98 | 0 | 0.03 | 0.54 | 0 | 0.18 |
| RB.AggPronSen_first_person | 0.13 | 0.06 | 0 | 0.22 | 0.56 | 0 | 0 |
| RB.AggPronSen_third_person | 0.12 | 0.38 | 0 | 0.02 | 0.69 | 0 | 0 |
| RB.AvgDepsSen_dobj | 0.1 | 0.07 | 0 | 0.07 | 0.31 | 0 | 0.46 |
| RB.AvgAdjectiveSen | 0.1 | 0.04 | 0 | 0.09 | 0.4 | 0 | 0.27 |
| RB.AvgDepsSen_cop | 0.1 | 0.14 | 0 | 0.04 | 0.59 | 0 | 0 |
| RB.AvgConnSen_reason_and_purpose | 0.09 | 0.05 | 0 | 0.06 | 0.25 | 0 | 0.46 |
| RB.AvgConnBl_conditions | 0.09 | 0.72 | 0 | 0.04 | 0.39 | 0 | 0 |
| RB.LangRhythmDiameter | 0.09 | 0.29 | 0 | 0.13 | 0.06 | 0.15 | 0 |
| RB.AvgDepsBl_acl | 0.09 | 0.74 | 0 | 0.03 | 0.39 | 0 | 0.09 |
| RB.AvgAOASen_Kuperman | 0.08 | 0.09 | 0 | 0.11 | 0.26 | 0 | 0.18 |
| RB.AvgConnBl_disjunctions | 0.08 | 0.52 | 0 | 0.03 | 0.21 | 0.08 | 0 |
| RB.AvgDepsSen_nmod | 0.08 | 0.02 | 0 | 0.15 | 0.17 | 0 | 0.27 |
| RB.AvgCommaBl | 0.08 | 0.78 | 0 | 0.02 | 0.36 | 0 | 0 |
| RB.AvgDepsBl_mwe | 0.07 | 0.67 | 0 | 0.01 | 0.33 | 0 | 0 |
| RB.AvgDepsBl_dep | 0.07 | 0.64 | 0 | 0.07 | 0.22 | 0 | 0.09 |
| RB.AvgConnSen_semi_coordinators | 0.06 | 0.13 | 0 | 0.01 | 0.01 | 0.11 | 0.27 |
| RB.AvgConnSen_conditions | 0.05 | 0.01 | 0 | 0.2 | 0 | 0 | 0 |
| RB.AvgConnBl_conjuncts | 0.04 | 0.42 | 0 | 0.01 | 0.14 | 0 | 0 |
| RB.LangRhythmId | 0.04 | 0.39 | 0 | 0.04 | 0.1 | 0 | 0 |
| RB.AvgDepsBl_csubj | 0.03 | 0.39 | 0 | 0 | 0.09 | 0 | 0 |
| RB.AvgDepsBl_iobj | 0.03 | 0.29 | 0 | 0.03 | 0.09 | 0 | 0 |
| RB.AvgDepsSen_nummod | 0.03 | 0.13 | 0 | 0.09 | 0.01 | 0 | 0 |
| RB.AvgDepsBl_auxpass | 0.03 | 0.36 | 0 | 0 | 0.11 | 0 | 0 |
| RB.AvgDepsBl_expl | 0.03 | 0.29 | 0 | 0 | 0.09 | 0.03 | 0 |
| RB.SenAsson | 0.03 | 0.37 | 0 | 0.02 | 0.1 | 0 | 0 |
| RB.AvgCommaSen | 0.03 | 0.14 | 0 | 0.05 | 0.02 | 0 | 0.18 |
| RB.AvgDepsSen_csubj | 0.01 | 0.04 | 0 | 0.02 | 0 | 0 | 0 |
| RB.AvgConnSen_disjunctions | 0.01 | 0.07 | 0 | 0.03 | 0.01 | 0 | 0 |
| RB.AvgDepsBl_parataxis | 0.01 | 0.2 | 0 | 0 | 0.03 | 0 | 0 |
| RB.AvgConnBl_complex_subordinators | 0 | 0.06 | 0 | 0 | 0.01 | 0 | 0 |
| RB.AvgConnSen_temporal_connectors | 0 | 0.01 | 0 | 0.01 | 0 | 0 | 0 |
Coh-Metrix Model 1
General Description
Model 1 has been replaced by the greatly simplified Model 2. Model 2 is recommended for current use.
Coh-Metrix Model 1 is an ensemble (formed by averaging predicted quality scores) of six sub-models that are detailed below.
All of these models used Coh-Metrix scores on 7 min narrative writing samples (“I once had a magic pencil and …”) from students in the fall, winter, and spring of Grades 2-5 (Mercer et al., 2019) to predict holistic writing quality on the samples (elo ratings calculated from paired comparisons). More details on the sample are available in (Mercer et al., 2019).
This scoring model was evaluated in the following publications: (Keller-Margulis et al., 2021; Matta et al., 2022)
Coh-Metrix Model 1a
This model was trained on fall Coh-Metrix scores from data described in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -10.8465 | 0.0266 | 0.1506 | 0.2663 | -0.0302 | 0.296 | 0.2609 | 0.136 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| DESWC | 19.42 | 42.06 | 5.86 | 4.76 | 26.22 | 7.84 | 45.19 | 26.59 |
| DESWLlt | 6.35 | 2.43 | 2.98 | 1.91 | 5.72 | 2.26 | 13.18 | 13.19 |
| LDMTLD | 4.88 | 8.17 | 4.01 | 3.11 | 6.4 | 4.13 | 4.19 | 10.99 |
| PCCONNp | 4.75 | 0.03 | 0 | 0.53 | 1.42 | 0.59 | 18.28 | 0 |
| PCNARp | 3.13 | 0 | 1.76 | 0.8 | 0 | 1.17 | 10.1 | 0 |
| WRDHYPn | 2.82 | 3.15 | 2.9 | 1.83 | 7.62 | 2.16 | 0 | 10.99 |
| PCVERBp | 2.35 | 0 | 0.99 | 0.6 | 0 | 1.23 | 7.35 | 0 |
| DESPL | 1.57 | 0.67 | 3.22 | 1.72 | 2.63 | 1.65 | 1.53 | 0 |
| SYNSTRUTa | 1.39 | 1.21 | 0.85 | 1.12 | 3.49 | 2.74 | 0 | 2.42 |
| PCDCp | 1.28 | 1.12 | 2 | 1.08 | 0 | 2.72 | 0 | 0.88 |
| DESWLsy | 1.26 | 0.48 | 2.09 | 1.23 | 1.3 | 1.34 | 0 | 3.08 |
| CNCTempx | 1.25 | 1.24 | 0.89 | 1.99 | 1.97 | 1.73 | 0 | 1.54 |
| LDTTRa | 1.25 | 0.48 | 2.01 | 0.89 | 2.69 | 1.65 | 0 | 3.08 |
| WRDFRQa | 1.23 | 1.38 | 1.86 | 1.58 | 1.86 | 0.82 | 0 | 3.08 |
| WRDVERB | 1.16 | 1.48 | 1.04 | 0.69 | 3.13 | 1.79 | 0 | 3.08 |
| LSASSpd | 1.11 | 0.1 | 1.95 | 1.43 | 3.46 | 1.39 | 0 | 1.54 |
| CNCTemp | 1.07 | 1.06 | 0.93 | 1.51 | 1.89 | 1.48 | 0 | 1.54 |
| CNCADC | 1 | 0.88 | 1.12 | 1.89 | 0 | 1.61 | 0 | 0 |
| SMINTEp | 0.99 | 1.86 | 0.75 | 1.33 | 3.22 | 1.3 | 0 | 1.54 |
| SMCAUSwn | 0.99 | 0.82 | 1.4 | 1.72 | 0 | 1.64 | 0 | 0 |
| DESWLsyd | 0.98 | 2.02 | 1.97 | 1.03 | 2.25 | 1.54 | 0 | 0.66 |
| CRFCWO1d | 0.97 | 1.03 | 1.67 | 1.5 | 0 | 1.64 | 0 | 0 |
| PCNARz | 0.95 | 0.74 | 1.29 | 0.88 | 3.68 | 1.5 | 0 | 1.54 |
| WRDHYPnv | 0.94 | 0.1 | 1.9 | 0.97 | 0 | 1.23 | 0 | 1.54 |
| WRDPRO | 0.94 | 1.26 | 1.77 | 1.28 | 0 | 1.68 | 0 | 0 |
| DESSLd | 0.93 | 0.15 | 1.67 | 1.09 | 0 | 1.79 | 0.18 | 0 |
| DESWLltd | 0.93 | 1.34 | 2.3 | 1.28 | 1.04 | 1.35 | 0 | 0 |
| DRPP | 0.93 | 0.99 | 2.18 | 1.12 | 0.47 | 1.62 | 0 | 0 |
| CNCLogic | 0.91 | 1.16 | 1.42 | 1.36 | 1.86 | 0.99 | 0 | 1.1 |
| LSAGN | 0.86 | 0.52 | 2.55 | 1.44 | 0 | 0.94 | 0 | 0 |
| PCCONNz | 0.83 | 1.35 | 1.09 | 1.09 | 0 | 1.21 | 0 | 0.88 |
| CRFCWOad | 0.82 | 0.32 | 1.51 | 1.44 | 0 | 1.24 | 0 | 0 |
| WRDADV | 0.81 | 0.07 | 1.3 | 1.24 | 0 | 1.51 | 0 | 0 |
| RDFRE | 0.81 | 0.86 | 1.29 | 0.98 | 2.81 | 1.65 | 0 | 0 |
| LSASS1d | 0.8 | 0.4 | 1.55 | 1.38 | 0.22 | 1.17 | 0 | 0 |
| WRDFRQmc | 0.8 | 1.22 | 1.35 | 0.32 | 0 | 1.92 | 0 | 0.66 |
| LDTTRc | 0.8 | 0.36 | 1.82 | 0.98 | 0.02 | 1.48 | 0 | 0 |
| PCVERBz | 0.8 | 0.11 | 1.55 | 0.87 | 0 | 0.93 | 0 | 1.54 |
| DRNP | 0.77 | 0.25 | 1.73 | 0.66 | 0 | 1.71 | 0 | 0 |
| WRDCNCc | 0.72 | 1.35 | 1.07 | 0.64 | 3.32 | 0 | 0 | 3.08 |
| CRFNOa | 0.72 | 0.4 | 0.74 | 1.33 | 0.63 | 1.25 | 0 | 0 |
| CNCPos | 0.71 | 0.33 | 0.88 | 1.02 | 0.09 | 0.64 | 0 | 1.54 |
| SYNMEDpos | 0.71 | 1.89 | 1.75 | 1.08 | 0 | 0.86 | 0 | 0 |
| LSAGNd | 0.68 | 0.78 | 1.5 | 1.1 | 0.83 | 0.94 | 0 | 0 |
| CNCCaus | 0.63 | 0.09 | 1.11 | 1.17 | 0 | 0.91 | 0 | 0 |
| DRVP | 0.63 | 1.02 | 0.82 | 0.6 | 0.03 | 0.7 | 0 | 1.54 |
| DRNEG | 0.63 | 0.4 | 0.76 | 1.13 | 0.03 | 1.09 | 0 | 0 |
| CRFCWOa | 0.61 | 0.32 | 0.46 | 1.25 | 0 | 0.98 | 0 | 0 |
| RDL2 | 0.61 | 0.3 | 0.31 | 0.87 | 0 | 1.45 | 0 | 0 |
| WRDPOLc | 0.61 | 0.56 | 0.85 | 1.57 | 0 | 0.5 | 0 | 0 |
| WRDIMGc | 0.61 | 0.19 | 0.14 | 0.74 | 0 | 0.87 | 0 | 1.54 |
| SMCAUSr | 0.6 | 0.32 | 1.58 | 0.23 | 0.02 | 1.5 | 0 | 0 |
| WRDFAMc | 0.6 | 1.32 | 1.09 | 0.86 | 1.25 | 0.96 | 0 | 0 |
| LSASSp | 0.59 | 0.14 | 0.79 | 1.05 | 0 | 0.99 | 0 | 0 |
| SMINTEr | 0.59 | 0.58 | 1.83 | 0.36 | 0 | 1.22 | 0 | 0 |
| SMCAUSlsa | 0.59 | 0.48 | 0.24 | 1.01 | 0.69 | 1.25 | 0 | 0 |
| WRDAOAc | 0.59 | 0.24 | 1.66 | 0.94 | 0.16 | 0.74 | 0 | 0 |
| SMCAUSvp | 0.54 | 0.02 | 0.93 | 1.33 | 0 | 0.46 | 0 | 0 |
| DRAP | 0.54 | 0.23 | 0.46 | 0.94 | 0 | 1.05 | 0 | 0 |
| WRDHYPv | 0.54 | 1.15 | 0.28 | 1.17 | 1.97 | 0.75 | 0 | 0 |
| PCTEMPp | 0.53 | 0.38 | 1.05 | 0.38 | 0 | 0.82 | 0 | 0.88 |
| SYNLE | 0.49 | 0.34 | 1.13 | 0.62 | 1.27 | 0.83 | 0 | 0 |
| PCDCz | 0.48 | 0 | 0 | 1.93 | 0 | 0 | 0 | 0 |
| CRFANPa | 0.46 | 0.18 | 0.76 | 0.77 | 0 | 0.78 | 0 | 0 |
| WRDNOUN | 0.44 | 0.54 | 0.56 | 0.53 | 0 | 0.97 | 0 | 0 |
| DESSC | 0.43 | 0 | 0 | 1.72 | 0 | 0 | 0 | 0 |
| CNCNeg | 0.43 | 0 | 0 | 1.74 | 0 | 0 | 0 | 0 |
| WRDPRP3s | 0.41 | 0.28 | 0.76 | 0.45 | 1.91 | 0.85 | 0 | 0 |
| PCREFp | 0.4 | 0 | 0.06 | 0.46 | 0 | 1.13 | 0 | 0 |
| PCREFz | 0.39 | 0.33 | 0.47 | 0.43 | 0 | 0.94 | 0 | 0 |
| WRDADJ | 0.38 | 0.08 | 0.99 | 0.59 | 0 | 0.51 | 0 | 0 |
| PCCNCz | 0.38 | 0.26 | 1.18 | 0.32 | 0 | 0.73 | 0 | 0 |
| CRFCWO1 | 0.37 | 0 | 0 | 1.5 | 0 | 0 | 0 | 0 |
| SMCAUSv | 0.35 | 0.38 | 0.39 | 0.53 | 0 | 0.68 | 0 | 0 |
| WRDMEAc | 0.34 | 1.04 | 0.21 | 0.57 | 2.34 | 0.56 | 0 | 0 |
| CNCAdd | 0.34 | 0 | 0 | 1.38 | 0 | 0 | 0 | 0 |
| WRDFRQc | 0.34 | 0.53 | 0.54 | 0.83 | 0 | 0.28 | 0 | 0 |
| PCCNCp | 0.32 | 0 | 0.57 | 0.13 | 0 | 0.93 | 0 | 0 |
| SYNSTRUTt | 0.32 | 0 | 0 | 1.28 | 0 | 0 | 0 | 0 |
| SYNNP | 0.32 | 0.14 | 0.03 | 0.26 | 0 | 1.02 | 0 | 0 |
| LSASS1 | 0.32 | 0 | 0 | 1.3 | 0 | 0 | 0 | 0 |
| CRFSOa | 0.31 | 0 | 0 | 1.25 | 0 | 0 | 0 | 0 |
| PCSYNp | 0.29 | 0.55 | 0.68 | 0 | 0.13 | 0.85 | 0 | 0 |
| SYNMEDlem | 0.28 | 0 | 0 | 1.12 | 0 | 0 | 0 | 0 |
| SYNMEDwrd | 0.26 | 0 | 0 | 1.03 | 0 | 0 | 0 | 0 |
| RDFKGL | 0.26 | 0 | 0 | 1.03 | 0 | 0 | 0 | 0 |
| WRDPRP3p | 0.25 | 0.01 | 0.84 | 0.02 | 0 | 0.66 | 0 | 0 |
| CNCAll | 0.22 | 0 | 0 | 0.91 | 0 | 0 | 0 | 0 |
| CRFAOa | 0.21 | 0 | 0 | 0.84 | 0 | 0 | 0 | 0 |
| PCTEMPz | 0.21 | 0 | 0 | 0.85 | 0 | 0 | 0 | 0 |
| DESSL | 0.15 | 0 | 0 | 0.59 | 0 | 0 | 0 | 0 |
| SMTEMP | 0.13 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 |
| CRFAO1 | 0.09 | 0 | 0 | 0.36 | 0 | 0 | 0 | 0 |
| CRFANP1 | 0.08 | 0 | 0 | 0.32 | 0 | 0 | 0 | 0 |
| PCSYNz | 0.04 | 0 | 0 | 0.17 | 0 | 0 | 0 | 0 |
| CRFSO1 | 0.03 | 0 | 0 | 0.14 | 0 | 0 | 0 | 0 |
| CRFNO1 | 0.02 | 0 | 0 | 0.08 | 0 | 0 | 0 | 0 |
Coh-Metrix Model 1b
This model used Coh-Metrix scores from 7 min narrative writing samples in winter (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -9.468 | 0.2532 | -0.0876 | 0.2097 | 0.0554 | 0.2458 | 0.2979 | 0.0974 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| DESWC | 20.79 | 30 | 7.75 | 3.44 | 27.38 | 7.68 | 37.87 | 27.6 |
| LDMTLD | 6.27 | 12.21 | 3.55 | 1.94 | 3.85 | 4.41 | 8.17 | 3.39 |
| DESWLlt | 6.11 | 3.24 | 1.99 | 1.08 | 6.03 | 2.23 | 14.98 | 13.8 |
| CRFCWO1d | 5.2 | 0.54 | 1.57 | 1.71 | 0.35 | 1.98 | 19.51 | 0 |
| LSAGN | 4.28 | 7.08 | 2.73 | 2.24 | 1.41 | 3.97 | 0 | 17.19 |
| RDL2 | 3.21 | 0.71 | 1.62 | 1.3 | 0 | 1.12 | 11.39 | 0 |
| DESPL | 2.56 | 0.19 | 3.82 | 1.84 | 6.29 | 2.53 | 5.18 | 0 |
| SYNLE | 1.48 | 3.13 | 0.87 | 1.41 | 1.45 | 2.12 | 0 | 0.48 |
| LSAGNd | 1.29 | 0.53 | 1.67 | 1.39 | 0 | 1.34 | 0 | 7.02 |
| WRDVERB | 1.28 | 1.66 | 1.91 | 1.32 | 3.02 | 2.19 | 0 | 0 |
| WRDPRP3s | 1.2 | 1.06 | 2.64 | 1.2 | 3.48 | 2.15 | 0 | 0 |
| RDFRE | 1.17 | 1.26 | 2.36 | 0.2 | 0 | 2.28 | 0 | 3.39 |
| WRDIMGc | 1.1 | 1.23 | 0.57 | 0.96 | 3.24 | 1.06 | 0 | 3.39 |
| DESSLd | 1.07 | 0.9 | 0.91 | 1.18 | 0 | 0.93 | 1.95 | 0 |
| LDTTRa | 1.06 | 0.96 | 3.04 | 0.83 | 0 | 1.12 | 0 | 3.39 |
| SYNMEDpos | 1.05 | 0.29 | 2.03 | 1.57 | 0 | 1.54 | 0 | 3.39 |
| WRDNOUN | 1.02 | 0.83 | 1.48 | 1.39 | 5.64 | 1.14 | 0 | 0 |
| PCCNCz | 1.01 | 0.78 | 1.56 | 1.26 | 0 | 1.22 | 0 | 3.39 |
| WRDHYPnv | 1.01 | 1.06 | 0.45 | 1.07 | 1.76 | 1.1 | 0 | 3.39 |
| DESWLltd | 0.95 | 1.21 | 0.88 | 1.2 | 2.76 | 1.52 | 0 | 0 |
| LSASSp | 0.91 | 0.91 | 1.57 | 1.34 | 0 | 1.92 | 0 | 0 |
| WRDHYPv | 0.88 | 0.72 | 1.87 | 1.14 | 2.79 | 1.32 | 0 | 0 |
| PCCNCp | 0.88 | 0 | 1.25 | 1.28 | 1.4 | 1.19 | 0 | 3.39 |
| SMCAUSwn | 0.87 | 1.08 | 1.35 | 1.6 | 2.18 | 0.78 | 0 | 0 |
| LSASS1d | 0.86 | 0.25 | 1.96 | 1.47 | 3.26 | 1.3 | 0 | 0 |
| CNCAdd | 0.84 | 0.74 | 1.13 | 0.8 | 1.85 | 0.53 | 0 | 3.39 |
| SYNNP | 0.82 | 1.25 | 1.32 | 0.72 | 4.14 | 0.68 | 0 | 0 |
| DESWLsy | 0.82 | 0.76 | 0.59 | 1.01 | 0 | 1.33 | 0.84 | 0 |
| DESWLsyd | 0.81 | 1.52 | 0.69 | 1.28 | 0 | 0.96 | 0.13 | 0 |
| WRDFRQmc | 0.75 | 0.84 | 0.92 | 0.99 | 2.76 | 1.03 | 0 | 0 |
| PCVERBz | 0.74 | 0.37 | 1.53 | 1.4 | 0 | 1.59 | 0 | 0 |
| PCREFp | 0.73 | 0 | 0.41 | 0.55 | 5.81 | 0.26 | 0 | 3.39 |
| CRFAOa | 0.72 | 0.22 | 1.88 | 1.32 | 0 | 1.6 | 0 | 0 |
| DRVP | 0.71 | 1.3 | 0.17 | 0.73 | 2.1 | 1.01 | 0 | 0 |
| CRFCWOad | 0.71 | 0.1 | 1.93 | 1.51 | 0 | 1.49 | 0 | 0 |
| SMCAUSv | 0.68 | 1.02 | 1.48 | 0.65 | 0 | 1.26 | 0 | 0 |
| PCSYNz | 0.68 | 0.41 | 1.89 | 0.69 | 0 | 1.78 | 0 | 0 |
| PCNARz | 0.67 | 0.59 | 1.31 | 1.29 | 0 | 1.15 | 0 | 0 |
| CRFCWO1 | 0.67 | 0.42 | 1.72 | 1.38 | 0 | 1.12 | 0 | 0 |
| CRFANPa | 0.66 | 0.22 | 1.25 | 1.25 | 0 | 1.57 | 0 | 0 |
| CRFNOa | 0.66 | 0.63 | 0.71 | 1.5 | 0 | 1.08 | 0 | 0 |
| DRPP | 0.66 | 0.69 | 1.38 | 0.81 | 2.9 | 0.69 | 0 | 0 |
| CNCTemp | 0.65 | 0.55 | 0.28 | 1.47 | 0.68 | 1.12 | 0 | 0 |
| SMINTEr | 0.63 | 0.72 | 1.23 | 0.55 | 0 | 1.56 | 0 | 0 |
| CNCNeg | 0.62 | 1.15 | 0.39 | 0.92 | 0 | 0.95 | 0 | 0 |
| LDTTRc | 0.62 | 0.65 | 1.34 | 1.03 | 0.22 | 0.99 | 0 | 0 |
| WRDADV | 0.62 | 0.62 | 0.79 | 0.93 | 0 | 1.4 | 0 | 0 |
| SMINTEp | 0.61 | 0.13 | 1.26 | 1.31 | 0 | 1.38 | 0 | 0 |
| WRDFRQa | 0.6 | 1.08 | 1.08 | 0.8 | 0 | 0.8 | 0 | 0 |
| SMCAUSlsa | 0.58 | 0.83 | 0.64 | 0.6 | 0 | 1.33 | 0 | 0 |
| SMCAUSvp | 0.58 | 0.33 | 1.47 | 1.12 | 0 | 1.08 | 0 | 0 |
| CNCAll | 0.58 | 0.92 | 1.12 | 0.75 | 1.02 | 0.64 | 0 | 0 |
| SYNSTRUTa | 0.56 | 0.4 | 1.51 | 0.94 | 0 | 1.03 | 0 | 0 |
| WRDADJ | 0.55 | 1.43 | 0.3 | 0.4 | 0.51 | 0.72 | 0 | 0 |
| WRDMEAc | 0.54 | 0.65 | 0.09 | 1.1 | 0 | 1.05 | 0 | 0 |
| DRNP | 0.53 | 0.53 | 1.42 | 0.45 | 0 | 1.25 | 0 | 0 |
| PCTEMPp | 0.52 | 0.51 | 1.21 | 0.8 | 0 | 0.95 | 0 | 0 |
| PCVERBp | 0.52 | 0 | 0.91 | 1.17 | 0 | 1.3 | 0 | 0 |
| SMCAUSr | 0.52 | 0.25 | 1.49 | 0.08 | 0 | 1.87 | 0 | 0 |
| PCSYNp | 0.51 | 0.01 | 1.27 | 0.44 | 0 | 1.82 | 0 | 0 |
| CNCCaus | 0.5 | 0.62 | 0.42 | 0.72 | 0 | 1.12 | 0 | 0 |
| WRDPRO | 0.49 | 0.65 | 0.75 | 0.93 | 0.14 | 0.62 | 0 | 0 |
| WRDAOAc | 0.48 | 0.84 | 0.97 | 0.55 | 0 | 0.7 | 0 | 0 |
| PCNARp | 0.46 | 0 | 1.14 | 1.07 | 0 | 0.96 | 0 | 0 |
| CNCTempx | 0.45 | 0.46 | 0.67 | 0.68 | 0 | 0.99 | 0 | 0 |
| WRDFRQc | 0.42 | 0.49 | 0.25 | 0.78 | 0 | 0.85 | 0 | 0 |
| PCCONNp | 0.39 | 0.8 | 0.87 | 0.29 | 0 | 0.57 | 0 | 0 |
| DRNEG | 0.37 | 0.17 | 0.47 | 0.85 | 0.01 | 0.75 | 0 | 0 |
| WRDPOLc | 0.36 | 0.19 | 0.72 | 0.73 | 0 | 0.72 | 0 | 0 |
| CNCLogic | 0.35 | 0.26 | 0.34 | 0.7 | 1.58 | 0.35 | 0 | 0 |
| DESSC | 0.34 | 0 | 0 | 1.84 | 0 | 0 | 0 | 0 |
| PCREFz | 0.33 | 0.51 | 0.84 | 0.55 | 0 | 0.3 | 0 | 0 |
| SYNMEDwrd | 0.32 | 0 | 0 | 1.72 | 0 | 0 | 0 | 0 |
| WRDFAMc | 0.31 | 0.46 | 0 | 0.71 | 0 | 0.42 | 0 | 0 |
| LSASSpd | 0.3 | 0 | 0 | 1.61 | 0 | 0 | 0 | 0 |
| PCDCp | 0.29 | 0.25 | 0.78 | 0.71 | 0 | 0.25 | 0 | 0 |
| SYNMEDlem | 0.29 | 0 | 0 | 1.57 | 0 | 0 | 0 | 0 |
| WRDHYPn | 0.29 | 0.36 | 1.3 | 0.66 | 0 | 0 | 0 | 0 |
| CRFSOa | 0.28 | 0 | 0 | 1.53 | 0 | 0 | 0 | 0 |
| DRAP | 0.26 | 0.3 | 0.38 | 0.69 | 0 | 0.21 | 0 | 0 |
| LSASS1 | 0.26 | 0 | 0 | 1.44 | 0 | 0 | 0 | 0 |
| CRFCWOa | 0.25 | 0 | 0 | 1.37 | 0 | 0 | 0 | 0 |
| WRDCNCc | 0.21 | 0 | 0 | 1.12 | 0 | 0 | 0 | 0 |
| SYNSTRUTt | 0.21 | 0 | 0 | 1.13 | 0 | 0 | 0 | 0 |
| PCTEMPz | 0.21 | 0 | 0 | 1.14 | 0 | 0 | 0 | 0 |
| SMTEMP | 0.19 | 0 | 0 | 1.02 | 0 | 0 | 0 | 0 |
| CRFANP1 | 0.19 | 0 | 0 | 1.02 | 0 | 0 | 0 | 0 |
| PCDCz | 0.18 | 0 | 0 | 0.99 | 0 | 0 | 0 | 0 |
| WRDPRP3p | 0.17 | 0 | 0.55 | 0 | 0 | 0.71 | 0 | 0 |
| CNCADC | 0.15 | 0 | 0 | 0.84 | 0 | 0 | 0 | 0 |
| CNCPos | 0.14 | 0 | 0 | 0.75 | 0 | 0 | 0 | 0 |
| PCCONNz | 0.12 | 0 | 0 | 0.65 | 0 | 0 | 0 | 0 |
| CRFAO1 | 0.1 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 |
| DESSL | 0.08 | 0 | 0 | 0.42 | 0 | 0 | 0 | 0 |
| RDFKGL | 0.06 | 0 | 0 | 0.32 | 0 | 0 | 0 | 0 |
| CRFSO1 | 0.04 | 0 | 0 | 0.2 | 0 | 0 | 0 | 0 |
| CRFNO1 | 0.02 | 0 | 0 | 0.08 | 0 | 0 | 0 | 0 |
Coh-Metrix Model 1c
This model used Coh-Metrix scores from 7 min narrative writing samples in spring (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -4.8423 | 0.5169 | 0.1348 | 0.6009 | -0.2375 | -0.4134 | 0.4001 | -0.0098 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| DESWC | 20.66 | 36.45 | 5.76 | 2.78 | 21.32 | 6.39 | 47.19 | 16.14 |
| WRDVERB | 5.51 | 3.32 | 1.88 | 1.07 | 0.58 | 2.06 | 22.99 | 2.79 |
| WRDHYPn | 4.27 | 2.98 | 2.38 | 1.13 | 3.13 | 1.75 | 14.74 | 1.28 |
| DESSLd | 3.23 | 1.17 | 0.87 | 1.74 | 3.05 | 2.13 | 10.33 | 0 |
| PCNARp | 2.65 | 2.58 | 2.72 | 1.77 | 10.29 | 2.17 | 0 | 4.99 |
| DESPL | 2.51 | 2.16 | 3.29 | 1.6 | 2.76 | 2.2 | 4.27 | 2.56 |
| DESWLltd | 1.8 | 4.36 | 1.75 | 1.09 | 1.39 | 1.51 | 0 | 6.97 |
| WRDNOUN | 1.59 | 1.69 | 2.13 | 0.8 | 5.92 | 1.42 | 0 | 5.81 |
| WRDFRQmc | 1.59 | 2.27 | 2.04 | 1.46 | 2.86 | 1.51 | 0 | 5.34 |
| DESWLlt | 1.55 | 0.7 | 2.05 | 0.77 | 6.89 | 1.97 | 0 | 3.72 |
| CRFANPa | 1.45 | 1.93 | 1.23 | 1.64 | 0.05 | 2.84 | 0 | 0 |
| LSASS1d | 1.43 | 2.3 | 1.5 | 1.82 | 0.08 | 1.92 | 0 | 0 |
| LDMTLD | 1.43 | 2.6 | 1.79 | 1.39 | 0 | 2.14 | 0 | 0 |
| CRFCWOa | 1.4 | 2.06 | 1.9 | 1.74 | 0 | 2.09 | 0 | 0 |
| WRDHYPv | 1.31 | 1.51 | 2.37 | 0.97 | 2.75 | 1.62 | 0 | 1.63 |
| PCDCz | 1.31 | 2.21 | 1.05 | 1.47 | 0 | 2 | 0 | 1.97 |
| WRDPRP3s | 1.28 | 1.96 | 1.36 | 0.66 | 3.27 | 1.43 | 0 | 0.81 |
| SMCAUSwn | 1.26 | 1.16 | 2.26 | 1.16 | 3.45 | 1.13 | 0 | 2.9 |
| SMCAUSvp | 1.25 | 2.1 | 0.97 | 1.55 | 0 | 1.8 | 0 | 0 |
| SYNSTRUTa | 1.17 | 1.47 | 1.74 | 1.81 | 0 | 1.38 | 0 | 3.72 |
| PCDCp | 1.16 | 0 | 1.81 | 1.28 | 3.29 | 2.05 | 0 | 4.53 |
| LSAGN | 1.12 | 0.71 | 1.72 | 1.81 | 0 | 2.08 | 0 | 2.09 |
| RDL2 | 1.04 | 0.86 | 2.17 | 0.93 | 2.07 | 1.45 | 0 | 1.28 |
| SMCAUSlsa | 1.01 | 0.63 | 1.59 | 1.07 | 3.13 | 0.94 | 0 | 3.72 |
| DRPP | 0.98 | 1.61 | 1.78 | 0.67 | 0.69 | 1.47 | 0 | 1.28 |
| LSAGNd | 0.97 | 0.08 | 2.29 | 2 | 0 | 1.63 | 0 | 0 |
| SYNMEDpos | 0.94 | 0.3 | 1.78 | 1.48 | 0 | 2.09 | 0 | 0.93 |
| CNCTemp | 0.9 | 1.06 | 0.85 | 0.91 | 1.61 | 1.18 | 0 | 0 |
| WRDADV | 0.89 | 1.56 | 1.9 | 0.67 | 0 | 1.4 | 0 | 0 |
| CNCPos | 0.87 | 0.61 | 0.82 | 0.74 | 1.97 | 1.58 | 0 | 2.44 |
| SMCAUSv | 0.86 | 1.16 | 0.91 | 1.27 | 0 | 1.22 | 0 | 0 |
| PCTEMPp | 0.85 | 0.19 | 1.68 | 1.09 | 2.36 | 0.96 | 0 | 2.09 |
| PCVERBz | 0.85 | 0.24 | 1.73 | 1.52 | 0 | 1.6 | 0 | 3.37 |
| LDTTRc | 0.84 | 1.39 | 1.26 | 0.82 | 0 | 1.37 | 0 | 0 |
| PCREFz | 0.78 | 0.25 | 1.2 | 0.56 | 2.5 | 1.41 | 0 | 0 |
| RDFKGL | 0.77 | 0.36 | 2.06 | 0.91 | 0 | 1.86 | 0 | 0 |
| LSASSp | 0.76 | 0.04 | 1.97 | 1.62 | 0 | 1.16 | 0 | 0.93 |
| PCVERBp | 0.75 | 0.3 | 1.09 | 1.28 | 0 | 1.55 | 0 | 0 |
| CRFCWO1d | 0.73 | 0.13 | 1.54 | 1.52 | 0 | 1.18 | 0 | 0 |
| DRNP | 0.7 | 0.54 | 1.62 | 0.83 | 0 | 1.51 | 0 | 0 |
| WRDPRO | 0.69 | 0.48 | 0.73 | 0.6 | 1.85 | 0.96 | 0 | 4.18 |
| SMCAUSr | 0.69 | 0.9 | 0.05 | 0.72 | 0.63 | 1.33 | 0 | 0 |
| WRDAOAc | 0.69 | 0.92 | 0.69 | 0.84 | 0.51 | 0.98 | 0 | 0 |
| LDTTRa | 0.68 | 0.05 | 2.31 | 0.95 | 0 | 1.57 | 0 | 0 |
| WRDCNCc | 0.67 | 0.53 | 1.3 | 0.38 | 3.63 | 0 | 0 | 1.28 |
| PCSYNz | 0.62 | 0.24 | 1.76 | 0.77 | 0 | 1.45 | 0 | 1.28 |
| CNCCaus | 0.61 | 0.72 | 0.73 | 0.73 | 0.12 | 1.1 | 0 | 0 |
| WRDMEAc | 0.57 | 0.51 | 0.68 | 0.45 | 2.11 | 0.46 | 0 | 0 |
| CNCTempx | 0.57 | 0.45 | 0.26 | 1.36 | 0.06 | 0.53 | 0 | 0 |
| DESWLsy | 0.56 | 0.34 | 0.85 | 0.55 | 0.35 | 0.89 | 0.49 | 1.28 |
| WRDPOLc | 0.56 | 0.48 | 0.89 | 0.63 | 0 | 1.31 | 0 | 0 |
| SYNLE | 0.55 | 0.35 | 0.09 | 0.62 | 0 | 1.67 | 0 | 0 |
| PCCNCz | 0.54 | 0.14 | 1.87 | 0.96 | 0 | 0.72 | 0 | 3.25 |
| DRNEG | 0.54 | 0.05 | 0.73 | 0.81 | 1.44 | 0.74 | 0 | 0 |
| WRDFRQa | 0.52 | 0.4 | 0.36 | 0.71 | 0 | 1.2 | 0 | 1.28 |
| WRDFRQc | 0.52 | 0.97 | 0.3 | 0.33 | 0 | 1.09 | 0 | 1.28 |
| DRVP | 0.5 | 0.11 | 0.79 | 0.77 | 0.61 | 0.88 | 0 | 0.81 |
| SYNNP | 0.5 | 0.21 | 0.65 | 0.88 | 0 | 1.04 | 0 | 0 |
| LSASSpd | 0.5 | 0 | 0 | 1.94 | 0 | 0 | 0 | 0 |
| CNCLogic | 0.49 | 0.55 | 0.41 | 0.68 | 0 | 0.9 | 0 | 0 |
| CNCADC | 0.49 | 0.38 | 1.61 | 0.46 | 0.24 | 0.93 | 0 | 0 |
| SYNSTRUTt | 0.48 | 0 | 0 | 1.83 | 0 | 0 | 0 | 0 |
| PCNARz | 0.46 | 0 | 0 | 1.77 | 0 | 0 | 0 | 0 |
| PCCNCp | 0.46 | 0 | 1.15 | 0.87 | 0 | 0.93 | 0 | 0 |
| WRDADJ | 0.45 | 0.29 | 1.12 | 0.72 | 0 | 0.76 | 0 | 0 |
| CRFCWOad | 0.44 | 0 | 0 | 1.69 | 0 | 0 | 0 | 0 |
| CRFAOa | 0.43 | 0 | 0 | 1.64 | 0 | 0 | 0 | 0 |
| PCSYNp | 0.43 | 0 | 1.45 | 0.62 | 0 | 1.01 | 0 | 0 |
| DESSC | 0.42 | 0 | 0 | 1.6 | 0 | 0 | 0 | 0 |
| CRFCWO1 | 0.42 | 0 | 0 | 1.6 | 0 | 0 | 0 | 0 |
| SYNMEDwrd | 0.42 | 0 | 0 | 1.61 | 0 | 0 | 0 | 0 |
| CRFAO1 | 0.41 | 0 | 0 | 1.55 | 0 | 0 | 0 | 0 |
| LSASS1 | 0.41 | 0 | 0 | 1.59 | 0 | 0 | 0 | 0 |
| SYNMEDlem | 0.41 | 0 | 0 | 1.59 | 0 | 0 | 0 | 0 |
| DRAP | 0.4 | 0.23 | 1.23 | 0.64 | 0 | 0.58 | 0 | 0 |
| SMINTEp | 0.38 | 0.36 | 1.06 | 0.68 | 0 | 0.29 | 0 | 0.81 |
| PCTEMPz | 0.38 | 0 | 0 | 1.45 | 0 | 0 | 0 | 0 |
| WRDPRP3p | 0.38 | 0.08 | 0.44 | 0.01 | 1.83 | 0.83 | 0 | 0 |
| SMTEMP | 0.37 | 0 | 0 | 1.44 | 0 | 0 | 0 | 0 |
| WRDHYPnv | 0.36 | 0.3 | 0.02 | 0.78 | 0 | 0.5 | 0 | 0 |
| CRFANP1 | 0.35 | 0 | 0 | 1.35 | 0 | 0 | 0 | 0 |
| CRFNOa | 0.34 | 0 | 0 | 1.32 | 0 | 0 | 0 | 0 |
| CRFSOa | 0.3 | 0 | 0 | 1.17 | 0 | 0 | 0 | 0 |
| PCREFp | 0.28 | 0 | 0.32 | 0.32 | 0 | 0.97 | 0 | 0 |
| DESWLsyd | 0.27 | 0.32 | 0.32 | 0.45 | 0 | 0.34 | 0 | 1.28 |
| PCCONNp | 0.26 | 0.07 | 1.18 | 0.05 | 0 | 0.87 | 0 | 0 |
| PCCONNz | 0.26 | 0.18 | 0.53 | 0.23 | 0 | 0.69 | 0 | 0 |
| DESSL | 0.24 | 0 | 0 | 0.91 | 0 | 0 | 0 | 0 |
| CRFNO1 | 0.22 | 0 | 0.98 | 0.06 | 0 | 0.81 | 0 | 0 |
| RDFRE | 0.21 | 0 | 0 | 0.79 | 0 | 0 | 0 | 0 |
| WRDFAMc | 0.21 | 0.29 | 0 | 0.07 | 1.19 | 0.02 | 0 | 0 |
| SMINTEr | 0.21 | 0.07 | 0.37 | 0.32 | 0 | 0.5 | 0 | 0 |
| CNCNeg | 0.15 | 0 | 0 | 0.58 | 0 | 0 | 0 | 0 |
| CNCAll | 0.12 | 0 | 0 | 0.46 | 0 | 0 | 0 | 0 |
| CNCAdd | 0.09 | 0 | 0 | 0.36 | 0 | 0 | 0 | 0 |
| WRDIMGc | 0.09 | 0 | 0 | 0.36 | 0 | 0 | 0 | 0 |
| CRFSO1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Coh-Metrix Model 1d
This model used principal components scores from 7 min narrative writing samples in fall (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -20.0773 | 0.0971 | 0.7558 | 0.5784 | -0.4401 | -4e-04 | 0.002 | 0.0227 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
PC1 = scores on 1st principal component extracted, …
Note: Importance is unavailable for support vector machines when PCA-based pre-processing is used (so all values for svm are 0).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| PC3 | 16.71 | 31.88 | 19.04 | 0 | 8.86 | 20.39 | 26.66 | 17.12 |
| PC5 | 12.28 | 18.78 | 13.53 | 0 | 8.6 | 14.58 | 19.92 | 8.98 |
| PC1 | 11.48 | 6.86 | 17.08 | 0 | 3.49 | 5.37 | 14.52 | 8.98 |
| PC8 | 7.3 | 4.81 | 7.77 | 0 | 7.07 | 4.77 | 9.88 | 8.47 |
| PC9 | 5 | 7.06 | 4.69 | 0 | 4.66 | 7.51 | 11.76 | 8.47 |
| PC4 | 4.73 | 2.76 | 5.98 | 0 | 3.09 | 2.6 | 0 | 7.97 |
| PC11 | 4.57 | 2.64 | 4.52 | 0 | 5.05 | 1.91 | 0 | 8.47 |
| PC7 | 2.91 | 4.92 | 2.9 | 0 | 2.16 | 5.93 | 0 | 8.47 |
| PC34 | 2.6 | 0.6 | 0.87 | 0 | 5.78 | 0.91 | 0 | 7.97 |
| PC14 | 2.37 | 0.66 | 2.18 | 0 | 3.18 | 0.56 | 0 | 2.2 |
| PC16 | 2.3 | 0.66 | 1.87 | 0 | 3.52 | 1.18 | 0 | 1.69 |
| PC21 | 2.27 | 0.65 | 1.5 | 0 | 3.75 | 1.41 | 0 | 6.78 |
| PC10 | 2.26 | 1.45 | 2.34 | 0 | 2.39 | 1.39 | 0 | 1.69 |
| PC30 | 2.07 | 0.38 | 0.88 | 0 | 4.61 | 1.45 | 0 | 0.51 |
| PC15 | 1.81 | 0 | 1.61 | 0 | 2.7 | 1.24 | 0 | 0.51 |
| PC31 | 1.8 | 1.2 | 0.69 | 0 | 3.86 | 1.42 | 0 | 1.19 |
| PC6 | 1.7 | 0.76 | 2.11 | 0 | 1.37 | 1.29 | 0 | 0 |
| PC35 | 1.7 | 0.15 | 0.51 | 0 | 4.07 | 1.11 | 5.51 | 0 |
| PC17 | 1.5 | 0.35 | 1.24 | 0 | 2.35 | 1.14 | 0 | 0 |
| PC12 | 1.4 | 0.88 | 1.45 | 0 | 1.59 | 0.23 | 0 | 0 |
| PC24 | 1.39 | 0.83 | 0.85 | 0 | 2.51 | 1.02 | 0 | 0 |
| PC13 | 1.3 | 0.62 | 1.24 | 0 | 1.66 | 0.98 | 0 | 0 |
| PC22 | 1.27 | 1.34 | 0.81 | 0 | 2.08 | 2.74 | 0 | 0 |
| PC19 | 1.11 | 2.25 | 0.72 | 0 | 1.51 | 4.51 | 0 | 0.51 |
| PC32 | 1.09 | 1.42 | 0.37 | 0 | 2.27 | 0.42 | 0 | 0 |
| PC2 | 0.89 | 1.16 | 1.19 | 0 | 0.33 | 2.11 | 0.87 | 0 |
| PC26 | 0.67 | 0.85 | 0.34 | 0 | 1.2 | 2.9 | 0 | 0 |
| PC33 | 0.64 | 0.61 | 0.17 | 0 | 1.32 | 0.89 | 7.71 | 0 |
| PC18 | 0.63 | 0.17 | 0.5 | 0 | 0.93 | 0.58 | 3.18 | 0 |
| PC29 | 0.6 | 0.71 | 0.24 | 0 | 1.18 | 2.26 | 0 | 0 |
| PC28 | 0.51 | 0.26 | 0.23 | 0 | 1.08 | 0.92 | 0 | 0 |
| PC20 | 0.43 | 0.87 | 0.27 | 0 | 0.63 | 0 | 0 | 0 |
| PC25 | 0.34 | 0.22 | 0.19 | 0 | 0.64 | 0.93 | 0 | 0 |
| PC27 | 0.33 | 0.86 | 0.12 | 0 | 0.52 | 2.07 | 0 | 0 |
| PC23 | 0.04 | 0.37 | 0 | 0 | 0 | 1.27 | 0 | 0 |
Proportion of Variance by Varimax Rotated Component (RC)
Due to space limitations, loadings for only the first ten principal components are displayed.
| Variable | RC2 | RC1 | RC4 | RC3 | RC8 | RC5 | RC6 | RC7 | RC10 | RC9 |
|---|---|---|---|---|---|---|---|---|---|---|
| SS loadings | 14.67 | 14.32 | 5.99 | 5.95 | 5.14 | 4.89 | 4.76 | 4.01 | 4.00 | 3.01 |
| Proportion Var | 0.15 | 0.15 | 0.06 | 0.06 | 0.05 | 0.05 | 0.05 | 0.04 | 0.04 | 0.03 |
| Cumulative Var | 0.15 | 0.30 | 0.36 | 0.42 | 0.47 | 0.53 | 0.57 | 0.62 | 0.66 | 0.69 |
| Proportion Explained | 0.22 | 0.21 | 0.09 | 0.09 | 0.08 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 |
| Cumulative Proportion | 0.22 | 0.43 | 0.52 | 0.61 | 0.69 | 0.76 | 0.83 | 0.90 | 0.95 | 1.00 |
Varimax Rotated Loadings
| Metric | RC2 | RC1 | RC4 | RC3 | RC8 | RC5 | RC6 | RC7 | RC10 | RC9 |
|---|---|---|---|---|---|---|---|---|---|---|
| DESSC | -0.01 | 0.8 | 0.31 | -0.06 | -0.06 | 0.02 | -0.04 | 0.05 | 0.29 | 0.01 |
| DESWC | 0.06 | 0.11 | 0.34 | 0.23 | -0.09 | -0.06 | -0.07 | 0.1 | 0.78 | -0.03 |
| DESPL | -0.01 | 0.8 | 0.31 | -0.06 | -0.06 | 0.02 | -0.04 | 0.05 | 0.29 | 0.01 |
| DESSL | -0.37 | -0.76 | -0.03 | 0.23 | 0.06 | 0.01 | -0.06 | 0.01 | 0.3 | 0.14 |
| DESSLd | 0.52 | -0.1 | 0.04 | 0.32 | 0.1 | -0.04 | -0.04 | -0.01 | 0.25 | -0.45 |
| DESWLsy | 0.08 | 0.09 | 0.71 | -0.01 | -0.06 | 0.24 | 0.25 | -0.07 | 0.02 | -0.02 |
| DESWLsyd | 0.08 | 0.07 | 0.66 | 0.01 | 0.01 | 0.22 | 0.18 | -0.13 | 0 | 0.13 |
| DESWLlt | 0.07 | 0.29 | 0.71 | 0.12 | 0.11 | 0.05 | 0.19 | 0.11 | 0.01 | -0.08 |
| DESWLltd | 0.22 | 0.24 | 0.69 | 0.08 | 0.06 | 0.21 | -0.09 | -0.06 | 0.02 | 0.2 |
| PCNARz | 0.74 | 0.23 | -0.02 | 0.1 | -0.22 | 0.02 | -0.51 | -0.1 | 0.01 | 0.07 |
| PCNARp | 0.61 | 0.35 | 0.13 | 0.06 | -0.2 | 0.07 | -0.48 | -0.03 | 0.06 | -0.01 |
| PCSYNz | -0.09 | 0.88 | 0.13 | -0.06 | -0.04 | -0.1 | 0.01 | 0.14 | -0.33 | 0.08 |
| PCSYNp | -0.19 | 0.84 | 0.16 | 0.01 | -0.01 | -0.04 | -0.03 | 0.15 | -0.22 | 0.12 |
| PCCNCz | -0.35 | -0.48 | -0.08 | 0.04 | 0.61 | -0.35 | 0.08 | 0.08 | -0.02 | 0.27 |
| PCCNCp | -0.12 | -0.35 | -0.02 | 0.03 | 0.57 | -0.38 | 0.1 | 0.14 | -0.08 | 0.25 |
| PCREFz | 0.71 | -0.34 | -0.25 | -0.08 | 0.02 | 0.04 | -0.03 | 0.14 | 0.07 | 0.48 |
| PCREFp | 0.45 | -0.36 | -0.26 | -0.03 | -0.03 | 0.04 | -0.1 | 0.13 | 0.16 | 0.52 |
| PCDCz | 0.12 | 0.17 | 0.12 | 0.9 | -0.09 | -0.15 | -0.02 | 0.22 | 0.04 | 0.04 |
| PCDCp | 0.12 | 0.13 | 0.14 | 0.85 | -0.14 | -0.14 | 0.02 | 0.16 | -0.01 | 0.08 |
| PCVERBz | -0.63 | -0.44 | -0.46 | 0.05 | -0.06 | 0.12 | 0.2 | 0.21 | 0.12 | 0.05 |
| PCVERBp | -0.47 | -0.21 | -0.5 | 0.06 | -0.01 | 0.13 | 0.21 | 0.3 | 0.11 | -0.08 |
| PCCONNz | 0.04 | 0.09 | 0.24 | 0.09 | -0.02 | 0.87 | -0.06 | -0.06 | 0.05 | 0.05 |
| PCCONNp | -0.08 | 0.01 | 0.02 | 0.01 | 0.01 | 0.81 | 0 | -0.1 | -0.12 | 0.01 |
| PCTEMPz | 0.67 | 0.64 | 0.17 | 0.06 | 0.06 | 0.01 | 0.01 | 0 | 0.01 | -0.28 |
| PCTEMPp | 0.4 | 0.37 | 0.04 | 0.04 | 0.17 | -0.16 | 0.01 | 0 | 0.14 | -0.35 |
| CRFNO1 | 0.61 | -0.16 | 0.11 | -0.12 | 0.06 | 0.06 | 0.49 | 0.2 | 0.17 | 0.09 |
| CRFAO1 | 0.88 | 0.21 | 0.13 | 0.02 | -0.01 | 0.08 | -0.06 | 0.09 | 0 | 0.08 |
| CRFSO1 | 0.64 | -0.13 | 0.12 | -0.06 | 0.04 | 0.01 | 0.51 | 0.18 | 0.19 | 0.01 |
| CRFNOa | 0.64 | -0.21 | 0.11 | -0.12 | 0.1 | 0.06 | 0.48 | 0.16 | 0.13 | 0.08 |
| CRFAOa | 0.91 | 0.21 | 0.07 | 0.06 | 0.06 | 0.06 | -0.09 | 0.01 | -0.07 | 0.06 |
| CRFSOa | 0.65 | -0.16 | 0.12 | -0.07 | 0.09 | 0.01 | 0.53 | 0.15 | 0.13 | 0.02 |
| CRFCWO1 | 0.88 | 0.18 | -0.08 | -0.03 | 0 | 0.07 | -0.06 | 0.14 | -0.01 | 0.2 |
| CRFCWO1d | 0.2 | 0.62 | 0.03 | 0.16 | 0.01 | 0.06 | -0.13 | -0.03 | 0.29 | 0.02 |
| CRFCWOa | 0.9 | 0.14 | -0.11 | -0.03 | 0.04 | 0.04 | -0.08 | 0.07 | -0.1 | 0.14 |
| CRFCWOad | 0.2 | 0.76 | -0.03 | 0.09 | 0.03 | 0.04 | 0.04 | -0.02 | 0.24 | -0.06 |
| CRFANP1 | 0.82 | 0.29 | 0.09 | 0.06 | 0 | 0.02 | -0.22 | -0.02 | -0.02 | 0 |
| CRFANPa | 0.85 | 0.09 | 0.01 | 0.12 | 0.05 | -0.03 | -0.25 | -0.06 | -0.13 | 0.04 |
| LSASS1 | 0.83 | 0.06 | 0.02 | -0.02 | 0.05 | -0.01 | 0.11 | -0.01 | 0.14 | -0.01 |
| LSASS1d | 0.16 | 0.7 | 0.09 | 0.05 | -0.01 | 0.05 | 0.08 | -0.05 | 0.26 | 0.06 |
| LSASSp | 0.85 | 0.01 | 0.03 | 0 | 0.08 | -0.01 | 0.11 | -0.04 | 0.08 | -0.04 |
| LSASSpd | 0.24 | 0.76 | 0.12 | 0.05 | 0 | 0.08 | 0.13 | 0.02 | 0.28 | 0.05 |
| LSAGN | 0.56 | 0.66 | 0.2 | -0.01 | -0.03 | 0 | 0.08 | 0.02 | 0.3 | -0.02 |
| LSAGNd | 0.78 | 0.41 | 0.12 | 0 | 0.08 | 0.03 | 0.07 | -0.02 | 0.08 | -0.15 |
| LDTTRc | -0.06 | -0.01 | 0.07 | -0.11 | 0.02 | 0.03 | -0.35 | -0.41 | -0.59 | 0.03 |
| LDTTRa | -0.16 | 0.12 | 0.12 | -0.07 | -0.11 | 0.11 | -0.04 | -0.17 | -0.76 | -0.16 |
| LDMTLD | -0.08 | 0.2 | 0.52 | 0.21 | -0.08 | 0.15 | -0.17 | 0.08 | 0.09 | -0.19 |
| CNCAll | -0.12 | -0.12 | -0.21 | 0.51 | 0.01 | -0.77 | 0.05 | -0.03 | -0.03 | 0.07 |
| CNCCaus | 0 | 0.01 | 0.08 | 0.86 | -0.02 | 0.08 | 0 | 0.06 | -0.09 | 0.01 |
| CNCLogic | 0.02 | 0 | 0.05 | 0.64 | -0.3 | -0.25 | -0.04 | 0.3 | 0.01 | 0.16 |
| CNCADC | 0.07 | 0.02 | 0.16 | 0.01 | -0.37 | -0.25 | -0.16 | 0.43 | 0.06 | 0.04 |
| CNCTemp | 0.01 | 0.11 | 0.09 | 0.31 | -0.1 | -0.28 | -0.01 | 0.11 | 0.03 | 0.22 |
| CNCTempx | 0.11 | 0.11 | -0.06 | 0.29 | 0.05 | 0.1 | 0.06 | 0 | 0.06 | 0.28 |
| CNCAdd | -0.11 | -0.19 | -0.33 | -0.04 | 0.09 | -0.83 | 0.05 | -0.08 | -0.06 | -0.04 |
| CNCPos | -0.13 | -0.1 | -0.22 | 0.53 | 0.12 | -0.67 | 0.09 | -0.14 | -0.06 | 0.09 |
| CNCNeg | 0.03 | 0.01 | 0.13 | 0.02 | -0.35 | -0.26 | -0.19 | 0.44 | 0.03 | 0.01 |
| SMCAUSv | -0.02 | 0.62 | 0.09 | 0.06 | 0.06 | -0.02 | -0.2 | 0.16 | -0.33 | 0.12 |
| SMCAUSvp | -0.02 | 0.44 | 0.11 | 0.52 | 0.02 | -0.01 | -0.17 | 0.2 | -0.32 | 0.05 |
| SMINTEp | 0.06 | 0.59 | 0.19 | 0.14 | 0.1 | -0.06 | -0.1 | 0.05 | -0.2 | 0.35 |
| SMCAUSr | -0.02 | -0.34 | 0 | 0.72 | -0.07 | 0.07 | 0.02 | 0.05 | 0.27 | -0.14 |
| SMINTEr | -0.07 | -0.31 | 0.01 | 0.72 | -0.05 | 0.1 | 0.06 | 0.01 | 0.26 | -0.13 |
| SMCAUSlsa | 0.03 | 0.03 | -0.29 | -0.15 | -0.03 | 0.34 | 0.35 | 0.48 | 0.1 | -0.02 |
| SMCAUSwn | 0.04 | 0.1 | -0.14 | 0.32 | 0.08 | 0.08 | 0.02 | 0.73 | 0.11 | 0.02 |
| SMTEMP | 0.67 | 0.64 | 0.17 | 0.04 | 0.07 | 0.02 | 0.01 | 0.01 | 0 | -0.26 |
| SYNLE | 0.01 | -0.19 | -0.02 | 0.34 | 0.03 | 0.02 | -0.06 | 0.07 | 0.34 | -0.19 |
| SYNNP | -0.05 | -0.04 | 0.03 | 0 | 0.14 | -0.09 | 0.61 | 0 | 0.1 | -0.32 |
| SYNMEDpos | 0.63 | 0.56 | 0.2 | 0.04 | 0.02 | 0.05 | 0.01 | -0.05 | -0.05 | -0.38 |
| SYNMEDwrd | 0.61 | 0.64 | 0.24 | 0.05 | 0.04 | 0.05 | -0.01 | -0.01 | -0.03 | -0.33 |
| SYNMEDlem | 0.61 | 0.63 | 0.24 | 0.05 | 0.04 | 0.03 | -0.02 | -0.01 | -0.02 | -0.34 |
| SYNSTRUTa | 0.06 | 0.8 | -0.01 | -0.05 | 0.08 | 0.19 | 0.1 | -0.06 | -0.16 | 0.1 |
| SYNSTRUTt | 0.08 | 0.83 | 0.03 | -0.03 | 0.05 | 0.2 | 0.07 | -0.08 | -0.2 | 0.08 |
| DRNP | 0.02 | -0.01 | -0.27 | -0.05 | 0.27 | 0.43 | 0.02 | 0.07 | -0.19 | 0.07 |
| DRVP | 0.15 | -0.03 | 0.14 | -0.08 | -0.09 | 0.18 | -0.63 | 0.14 | 0.02 | -0.02 |
| DRAP | -0.11 | 0.06 | 0.26 | 0.12 | -0.57 | -0.19 | 0.03 | -0.01 | -0.16 | 0.23 |
| DRPP | 0.13 | 0.23 | 0.27 | 0.09 | 0.2 | 0.09 | 0.04 | -0.16 | 0.27 | 0.25 |
| DRNEG | -0.13 | -0.23 | 0.1 | -0.1 | -0.33 | -0.01 | -0.18 | -0.01 | 0.28 | 0.05 |
| WRDNOUN | -0.15 | 0.11 | 0.07 | -0.07 | 0.29 | 0.09 | 0.7 | -0.09 | -0.09 | -0.03 |
| WRDVERB | -0.05 | -0.05 | 0.03 | -0.05 | 0.13 | -0.1 | -0.54 | 0.18 | 0.12 | -0.12 |
| WRDADJ | -0.01 | -0.04 | -0.13 | 0.01 | -0.11 | 0.1 | 0.33 | 0.01 | -0.16 | -0.5 |
| WRDADV | -0.11 | 0.01 | 0.29 | 0.2 | -0.7 | -0.18 | -0.04 | 0.02 | -0.01 | 0.17 |
| WRDPRO | 0.13 | -0.08 | -0.25 | -0.09 | -0.1 | 0.16 | -0.6 | -0.03 | -0.21 | 0.13 |
| WRDPRP3s | -0.14 | -0.02 | -0.1 | 0.13 | -0.01 | 0.02 | -0.17 | 0.12 | 0.05 | -0.17 |
| WRDPRP3p | -0.06 | 0.04 | 0.12 | 0.25 | 0.04 | -0.08 | 0 | -0.01 | 0.13 | -0.1 |
| WRDFRQc | -0.1 | -0.09 | -0.49 | 0.07 | -0.54 | 0.25 | -0.02 | -0.07 | 0.09 | 0.11 |
| WRDFRQa | -0.11 | -0.21 | -0.59 | -0.13 | -0.21 | -0.01 | 0.1 | -0.44 | 0.07 | 0.04 |
| WRDFRQmc | 0.09 | 0.64 | 0.01 | 0.13 | -0.03 | 0.07 | -0.16 | 0.25 | 0 | -0.08 |
| WRDAOAc | 0.1 | -0.02 | 0.39 | 0.12 | 0.03 | 0 | -0.19 | 0.27 | 0.11 | -0.04 |
| WRDFAMc | 0 | 0.02 | -0.28 | 0.15 | -0.19 | 0.19 | 0.02 | -0.15 | 0.31 | 0.18 |
| WRDCNCc | 0.09 | 0.1 | 0.24 | -0.08 | 0.77 | -0.05 | 0.11 | 0.15 | 0.14 | 0.15 |
| WRDIMGc | 0.09 | 0.04 | 0.22 | -0.14 | 0.86 | -0.03 | 0.12 | 0.06 | 0.03 | 0.02 |
| WRDMEAc | 0.06 | 0.04 | 0.22 | 0.03 | 0.73 | -0.04 | 0.03 | -0.06 | -0.09 | 0.06 |
| WRDPOLc | -0.01 | 0.08 | -0.23 | 0.32 | -0.03 | -0.03 | -0.13 | 0.66 | -0.04 | -0.05 |
| WRDHYPn | 0.12 | 0.15 | 0.38 | 0.13 | 0.13 | 0 | -0.1 | 0.66 | 0.13 | -0.02 |
| WRDHYPv | 0.01 | -0.01 | 0.02 | 0.14 | 0.27 | -0.19 | -0.22 | 0.39 | -0.03 | 0.26 |
| WRDHYPnv | 0.01 | 0.08 | 0.37 | 0.05 | 0.38 | 0.07 | 0.22 | 0.6 | 0.09 | 0.06 |
| RDFRE | 0.35 | 0.74 | -0.17 | -0.23 | -0.05 | -0.08 | -0.02 | 0.02 | -0.29 | -0.13 |
| RDFKGL | -0.36 | -0.76 | 0.04 | 0.23 | 0.06 | 0.03 | -0.03 | 0.01 | 0.3 | 0.14 |
| RDL2 | 0.5 | 0.5 | -0.34 | -0.01 | -0.27 | 0.29 | 0.01 | 0.01 | -0.04 | 0.24 |
Coh-Metrix Model 1e
This model used principal components scores from 7 min narrative writing samples in winter (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -10.9566 | 0.338 | 0.385 | 0.5608 | -0.2444 | 0.0443 | 0.0047 | -0.0256 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
PC1 = scores on 1st principal component extracted, …
Note: Importance is unavailable for support vector machines when PCA-based pre-processing is used (so all values for svm are 0).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| PC1 | 15.14 | 27.79 | 9.8 | 0 | 4.09 | 18.64 | 24.42 | 21.1 |
| PC4 | 9.3 | 10.16 | 11.41 | 0 | 4.87 | 7.73 | 13.26 | 10.85 |
| PC7 | 8.4 | 8.15 | 9.97 | 0 | 6.48 | 6.21 | 19.43 | 10.26 |
| PC11 | 6.67 | 7.23 | 6.54 | 0 | 5.69 | 5.83 | 11.19 | 10.26 |
| PC9 | 6.08 | 3.67 | 9.2 | 0 | 6.76 | 2 | 1.16 | 1.97 |
| PC10 | 5.49 | 3.61 | 7.03 | 0 | 5.42 | 3.37 | 8.76 | 9.47 |
| PC28 | 4.63 | 2.51 | 3.16 | 0 | 9.22 | 3.54 | 0 | 10.26 |
| PC16 | 3.93 | 2.03 | 4.15 | 0 | 5.33 | 2.65 | 0 | 10.26 |
| PC12 | 3.92 | 2.11 | 4.88 | 0 | 4.59 | 1.8 | 0 | 8.88 |
| PC33 | 3.09 | 2.27 | 1.71 | 0 | 7.03 | 2.57 | 0 | 1.97 |
| PC17 | 2.46 | 1.81 | 2.53 | 0 | 3.42 | 2.91 | 0 | 0.59 |
| PC31 | 2.32 | 2.4 | 1.32 | 0 | 4.7 | 1.59 | 0 | 0 |
| PC20 | 2.3 | 4.01 | 1.31 | 0 | 1.99 | 2.63 | 0 | 0 |
| PC18 | 2.29 | 1.13 | 2.55 | 0 | 3.68 | 2.3 | 16.31 | 0.59 |
| PC14 | 2.21 | 2.99 | 1.75 | 0 | 1.83 | 3.19 | 0 | 0.79 |
| PC22 | 2.1 | 0.93 | 2.13 | 0 | 3.83 | 2.78 | 0 | 0 |
| PC25 | 2.05 | 0.96 | 2.05 | 0 | 4.55 | 0 | 0 | 1.38 |
| PC19 | 2.02 | 1.4 | 2.05 | 0 | 2.95 | 2.31 | 0 | 0.99 |
| PC6 | 1.88 | 2.76 | 1.76 | 0 | 0.89 | 2.87 | 0 | 0 |
| PC3 | 1.79 | 0.55 | 3.72 | 0 | 0.94 | 1 | 0 | 0 |
| PC2 | 1.49 | 0.77 | 2.52 | 0 | 0.49 | 2.87 | 0.24 | 0 |
| PC15 | 1.38 | 0 | 2.17 | 0 | 2.43 | 0.98 | 0 | 0 |
| PC8 | 1.38 | 1.43 | 1.51 | 0 | 0.9 | 2.69 | 0 | 0 |
| PC13 | 1.31 | 1.74 | 1.23 | 0 | 1.17 | 1.25 | 0 | 0 |
| PC26 | 1.13 | 1.03 | 0.71 | 0 | 1.66 | 2.56 | 0 | 0 |
| PC32 | 1.03 | 2.12 | 0.2 | 0 | 0.73 | 2.06 | 0 | 0 |
| PC29 | 0.93 | 0.43 | 0.75 | 0 | 2.25 | 0.69 | 0 | 0 |
| PC34 | 0.68 | 1.1 | 0.12 | 0 | 0.53 | 2.38 | 0 | 0 |
| PC21 | 0.58 | 0.44 | 0.44 | 0 | 0.59 | 1.47 | 5.24 | 0.39 |
| PC5 | 0.5 | 0.16 | 0.73 | 0 | 0.16 | 1.73 | 0 | 0 |
| PC27 | 0.44 | 0.88 | 0.15 | 0 | 0.22 | 0.95 | 0 | 0 |
| PC23 | 0.37 | 0.46 | 0.17 | 0 | 0.16 | 1.67 | 0 | 0 |
| PC24 | 0.36 | 0.15 | 0.28 | 0 | 0.43 | 1.4 | 0 | 0 |
| PC30 | 0.36 | 0.82 | 0 | 0 | 0 | 1.41 | 0 | 0 |
Proportion of Variance by Varimax Rotated Component (RC)
Due to space limitations, loadings for only the first ten principal components are displayed.
| Variable | RC1 | RC3 | RC2 | RC4 | RC5 | RC6 | RC7 | RC10 | RC9 | RC8 |
|---|---|---|---|---|---|---|---|---|---|---|
| SS loadings | 17.16 | 14.47 | 6.31 | 5.42 | 5.10 | 5.09 | 4.07 | 3.84 | 3.40 | 3.20 |
| Proportion Var | 0.18 | 0.15 | 0.07 | 0.06 | 0.05 | 0.05 | 0.04 | 0.04 | 0.04 | 0.03 |
| Cumulative Var | 0.18 | 0.33 | 0.39 | 0.45 | 0.50 | 0.55 | 0.59 | 0.63 | 0.67 | 0.70 |
| Proportion Explained | 0.25 | 0.21 | 0.09 | 0.08 | 0.07 | 0.07 | 0.06 | 0.06 | 0.05 | 0.05 |
| Cumulative Proportion | 0.25 | 0.46 | 0.56 | 0.64 | 0.71 | 0.79 | 0.85 | 0.90 | 0.95 | 1.00 |
Varimax Rotated Loadings
| Metric | RC1 | RC3 | RC2 | RC4 | RC5 | RC6 | RC7 | RC10 | RC9 | RC8 |
|---|---|---|---|---|---|---|---|---|---|---|
| DESSC | 0.86 | -0.02 | 0.02 | 0.05 | -0.06 | 0.01 | 0.08 | 0.07 | 0.18 | 0.11 |
| DESWC | 0.18 | 0.16 | -0.09 | 0.21 | 0.05 | -0.09 | 0.19 | 0.21 | 0.63 | 0.38 |
| DESPL | 0.86 | -0.02 | 0.02 | 0.05 | -0.06 | 0.01 | 0.08 | 0.07 | 0.18 | 0.11 |
| DESSL | -0.81 | -0.25 | -0.04 | 0.15 | 0 | -0.07 | 0.1 | 0.12 | 0.34 | 0.01 |
| DESSLd | -0.04 | 0.46 | -0.08 | 0.14 | 0.1 | -0.15 | 0.07 | 0.17 | 0.01 | 0.51 |
| DESWLsy | 0.14 | 0.21 | 0.02 | -0.1 | -0.16 | 0.84 | -0.05 | -0.06 | -0.06 | -0.05 |
| DESWLsyd | 0.13 | 0.17 | -0.07 | -0.04 | -0.14 | 0.79 | -0.08 | 0.03 | -0.14 | -0.04 |
| DESWLlt | 0.22 | 0.12 | 0.21 | -0.04 | -0.01 | 0.83 | -0.1 | 0.02 | 0.02 | -0.05 |
| DESWLltd | 0.18 | 0.13 | -0.05 | -0.07 | -0.17 | 0.67 | -0.11 | 0.11 | -0.13 | -0.09 |
| PCNARz | 0.33 | 0.64 | -0.37 | 0.18 | 0 | -0.25 | 0.34 | 0.28 | -0.03 | 0.05 |
| PCNARp | 0.42 | 0.53 | -0.34 | 0.22 | -0.04 | -0.17 | 0.23 | 0.28 | 0 | 0.04 |
| PCSYNz | 0.9 | -0.07 | 0.06 | -0.01 | 0.11 | 0.14 | -0.05 | -0.09 | -0.14 | -0.18 |
| PCSYNp | 0.85 | -0.16 | 0.07 | 0.14 | 0.08 | 0.17 | -0.04 | -0.07 | 0.01 | -0.15 |
| PCCNCz | -0.56 | -0.3 | 0.65 | -0.14 | 0.17 | -0.11 | -0.1 | 0.03 | -0.04 | -0.19 |
| PCCNCp | -0.42 | -0.11 | 0.69 | 0.05 | 0.09 | -0.03 | -0.06 | 0.13 | 0.09 | -0.16 |
| PCREFz | -0.36 | 0.73 | -0.06 | -0.07 | -0.02 | -0.19 | -0.04 | 0.08 | 0.16 | -0.46 |
| PCREFp | -0.42 | 0.45 | -0.14 | -0.17 | -0.08 | -0.26 | 0 | 0.07 | 0.18 | -0.46 |
| PCDCz | 0.14 | 0.24 | -0.15 | 0.86 | 0.18 | 0.01 | 0.13 | 0.04 | 0.15 | 0.06 |
| PCDCp | 0.19 | 0.2 | -0.14 | 0.82 | 0.21 | 0.02 | 0.09 | -0.01 | 0.09 | 0.03 |
| PCVERBz | -0.56 | -0.65 | -0.12 | 0.01 | -0.04 | -0.12 | -0.39 | 0.14 | 0.05 | 0 |
| PCVERBp | -0.33 | -0.56 | -0.15 | 0.09 | 0.05 | -0.14 | -0.51 | 0.17 | -0.07 | 0.13 |
| PCCONNz | 0.05 | -0.04 | -0.1 | 0.13 | -0.93 | 0.06 | -0.11 | -0.02 | 0.07 | -0.03 |
| PCCONNp | 0.04 | -0.03 | -0.02 | -0.08 | -0.75 | 0 | -0.13 | -0.09 | -0.13 | -0.04 |
| PCTEMPz | 0.68 | 0.62 | -0.03 | 0.12 | 0.05 | 0.15 | 0 | 0.11 | -0.07 | 0.25 |
| PCTEMPp | 0.42 | 0.46 | 0.13 | 0.08 | 0.04 | 0.03 | 0.02 | 0.21 | -0.11 | 0.17 |
| CRFNO1 | -0.11 | 0.73 | 0.26 | 0.11 | -0.01 | 0.12 | -0.16 | -0.08 | 0.1 | 0.08 |
| CRFAO1 | 0.33 | 0.86 | -0.07 | 0.05 | 0.01 | 0.1 | 0.01 | 0.09 | -0.03 | -0.04 |
| CRFSO1 | -0.1 | 0.77 | 0.27 | 0.09 | -0.03 | 0.16 | -0.1 | -0.12 | 0.12 | 0.14 |
| CRFNOa | -0.16 | 0.77 | 0.25 | 0.07 | 0.05 | 0.07 | -0.12 | -0.07 | 0.13 | 0.12 |
| CRFAOa | 0.31 | 0.88 | -0.08 | 0.03 | 0.01 | 0.1 | 0.03 | 0.1 | -0.06 | -0.01 |
| CRFSOa | -0.14 | 0.78 | 0.25 | 0.08 | 0.02 | 0.13 | -0.1 | -0.11 | 0.11 | 0.14 |
| CRFCWO1 | 0.24 | 0.86 | -0.11 | 0.03 | 0.05 | 0.05 | -0.05 | 0.08 | 0 | -0.19 |
| CRFCWO1d | 0.8 | 0.14 | -0.04 | 0.01 | -0.06 | 0.1 | 0.01 | 0.07 | 0.14 | 0.03 |
| CRFCWOa | 0.21 | 0.89 | -0.14 | 0.01 | 0.06 | 0.01 | -0.02 | 0.09 | -0.01 | -0.16 |
| CRFCWOad | 0.82 | 0.2 | -0.03 | -0.02 | -0.07 | 0.02 | 0.05 | 0.09 | 0.17 | 0.13 |
| CRFANP1 | 0.47 | 0.66 | -0.19 | 0.05 | 0.05 | 0.06 | 0.02 | 0.18 | -0.04 | -0.07 |
| CRFANPa | 0.28 | 0.71 | -0.21 | 0.03 | 0.04 | 0.11 | 0.03 | 0.18 | -0.16 | -0.07 |
| LSASS1 | 0.22 | 0.83 | 0.09 | 0 | -0.01 | 0.04 | 0.03 | 0.04 | 0.17 | 0.03 |
| LSASS1d | 0.71 | 0.21 | 0.01 | -0.04 | -0.09 | 0.03 | 0.04 | 0.08 | 0.26 | 0.14 |
| LSASSp | 0.17 | 0.87 | 0.06 | -0.01 | 0.04 | 0 | 0.06 | 0 | 0.17 | 0.02 |
| LSASSpd | 0.72 | 0.28 | 0.04 | -0.08 | -0.09 | 0.07 | 0.06 | 0.05 | 0.32 | 0.1 |
| LSAGN | 0.75 | 0.52 | 0.02 | -0.01 | -0.02 | 0.04 | 0.1 | 0.06 | 0.28 | 0.06 |
| LSAGNd | 0.48 | 0.78 | 0.02 | 0.03 | 0.03 | 0.05 | 0.06 | 0.05 | 0.16 | 0.14 |
| LDTTRc | -0.07 | -0.35 | 0.08 | -0.11 | -0.11 | 0.09 | 0.1 | 0.03 | -0.74 | 0.19 |
| LDTTRa | 0.05 | -0.3 | 0.3 | -0.02 | -0.14 | 0.4 | 0.06 | -0.1 | -0.66 | 0.04 |
| LDMTLD | 0.24 | -0.14 | 0.17 | 0.28 | -0.11 | 0.51 | 0.27 | 0.03 | 0.02 | 0.41 |
| CNCAll | -0.12 | 0.07 | -0.06 | 0.32 | 0.86 | -0.16 | 0.08 | -0.15 | 0.02 | -0.07 |
| CNCCaus | 0.03 | 0.08 | 0.03 | 0.84 | -0.1 | -0.02 | -0.14 | 0.04 | -0.06 | 0.06 |
| CNCLogic | -0.07 | -0.09 | -0.31 | 0.62 | 0.35 | -0.01 | 0.33 | -0.07 | 0.19 | -0.13 |
| CNCADC | -0.04 | -0.12 | -0.05 | 0.1 | 0.36 | 0.13 | 0.52 | 0.33 | -0.02 | 0.06 |
| CNCTemp | 0.14 | 0.09 | -0.31 | 0.08 | 0.34 | -0.01 | 0.32 | -0.21 | 0.2 | -0.23 |
| CNCTempx | 0.15 | -0.11 | 0.03 | 0.39 | 0.07 | 0.02 | 0.05 | -0.02 | -0.22 | -0.1 |
| CNCAdd | -0.14 | 0.01 | 0.04 | -0.19 | 0.89 | -0.14 | -0.04 | -0.08 | -0.04 | -0.01 |
| CNCPos | -0.08 | 0.11 | -0.05 | 0.33 | 0.73 | -0.2 | -0.16 | -0.26 | 0 | -0.1 |
| CNCNeg | -0.07 | -0.13 | -0.11 | 0.03 | 0.37 | 0.11 | 0.55 | 0.31 | 0.02 | 0.01 |
| SMCAUSv | 0.64 | 0.04 | 0.07 | 0.05 | -0.09 | 0.14 | -0.05 | 0.06 | -0.15 | -0.33 |
| SMCAUSvp | 0.49 | 0.12 | 0.11 | 0.55 | -0.14 | 0.06 | -0.03 | 0.09 | -0.06 | -0.17 |
| SMINTEp | 0.66 | 0.08 | 0.13 | 0.05 | -0.18 | 0.11 | 0.09 | 0.03 | 0.12 | -0.27 |
| SMCAUSr | -0.36 | 0.16 | -0.05 | 0.58 | -0.13 | -0.13 | 0.05 | 0.08 | 0.18 | 0.3 |
| SMINTEr | -0.43 | 0.12 | -0.08 | 0.68 | -0.06 | -0.07 | -0.06 | 0.04 | 0.12 | 0.25 |
| SMCAUSlsa | -0.01 | 0.06 | -0.17 | 0.13 | 0.12 | 0.04 | -0.54 | 0.07 | 0.29 | -0.01 |
| SMCAUSwn | 0.28 | 0.09 | -0.01 | 0.23 | 0.02 | 0.22 | -0.35 | 0.54 | 0.22 | 0.22 |
| SMTEMP | 0.69 | 0.62 | -0.03 | 0.11 | 0.04 | 0.15 | -0.01 | 0.11 | -0.07 | 0.24 |
| SYNLE | -0.13 | -0.11 | -0.1 | -0.14 | 0.05 | 0.15 | 0.11 | 0.03 | 0.18 | -0.06 |
| SYNNP | 0 | -0.05 | 0.29 | -0.17 | 0 | 0.38 | -0.21 | -0.5 | 0.09 | 0.35 |
| SYNMEDpos | 0.66 | 0.51 | -0.06 | 0.11 | 0.04 | 0.13 | 0.05 | 0.1 | -0.11 | 0.4 |
| SYNMEDwrd | 0.71 | 0.53 | -0.04 | 0.12 | 0.03 | 0.17 | 0.01 | 0.09 | -0.07 | 0.35 |
| SYNMEDlem | 0.7 | 0.54 | -0.04 | 0.12 | 0.03 | 0.16 | 0.02 | 0.09 | -0.08 | 0.35 |
| SYNSTRUTa | 0.81 | 0.14 | -0.08 | 0.08 | -0.12 | 0.17 | -0.06 | 0.05 | -0.05 | -0.13 |
| SYNSTRUTt | 0.85 | 0.15 | -0.05 | 0.06 | -0.12 | 0.16 | -0.05 | 0.02 | -0.04 | -0.15 |
| DRNP | -0.04 | -0.1 | 0.15 | 0.02 | -0.43 | -0.03 | -0.26 | -0.02 | -0.12 | -0.28 |
| DRVP | -0.05 | 0.07 | -0.08 | -0.14 | -0.05 | -0.14 | 0 | 0.65 | 0 | -0.02 |
| DRAP | 0.09 | 0 | -0.21 | 0.22 | 0.26 | -0.09 | 0.42 | -0.01 | 0.06 | -0.36 |
| DRPP | 0.11 | -0.02 | 0.05 | 0.09 | -0.01 | 0.06 | -0.45 | -0.03 | 0 | 0.05 |
| DRNEG | 0.03 | 0.03 | -0.14 | 0.08 | 0.1 | -0.15 | 0.53 | -0.11 | 0.03 | 0.09 |
| WRDNOUN | 0.04 | -0.06 | 0.49 | -0.12 | -0.16 | 0.28 | -0.41 | -0.4 | 0.02 | 0.06 |
| WRDVERB | 0.17 | 0.17 | 0.05 | -0.13 | -0.06 | 0 | 0.17 | 0.65 | -0.12 | 0.03 |
| WRDADJ | 0.05 | 0.01 | -0.09 | 0.04 | -0.06 | 0.37 | 0.01 | -0.26 | -0.03 | 0.29 |
| WRDADV | 0.04 | 0.04 | -0.24 | 0.38 | 0.29 | 0 | 0.47 | -0.08 | 0.22 | -0.19 |
| WRDPRO | -0.11 | 0.01 | -0.27 | -0.03 | -0.23 | -0.54 | 0.37 | 0.2 | -0.13 | -0.19 |
| WRDPRP3s | 0.05 | 0.01 | -0.09 | 0.14 | -0.03 | -0.03 | 0.45 | -0.03 | 0.1 | 0.22 |
| WRDPRP3p | -0.23 | -0.17 | -0.12 | 0.23 | 0.04 | 0.13 | -0.02 | 0.04 | 0.19 | 0.04 |
| WRDFRQc | -0.12 | -0.25 | -0.58 | 0.26 | -0.15 | -0.24 | -0.05 | 0.31 | 0.05 | 0.06 |
| WRDFRQa | -0.23 | -0.19 | -0.49 | -0.11 | 0.06 | -0.47 | -0.23 | -0.06 | -0.26 | 0.07 |
| WRDFRQmc | 0.66 | -0.02 | -0.21 | 0.05 | 0.11 | 0.1 | -0.08 | 0.15 | -0.07 | 0.18 |
| WRDAOAc | 0.12 | 0.14 | 0.17 | 0.16 | 0.04 | 0.32 | 0.03 | -0.09 | 0.07 | 0.13 |
| WRDFAMc | -0.09 | -0.12 | -0.47 | 0.22 | -0.18 | -0.23 | -0.08 | 0.22 | 0.12 | -0.06 |
| WRDCNCc | 0.03 | 0.02 | 0.83 | -0.12 | 0.04 | -0.03 | -0.06 | 0.02 | -0.09 | 0.05 |
| WRDIMGc | 0.08 | 0.07 | 0.88 | -0.06 | -0.05 | 0 | -0.11 | -0.02 | -0.15 | 0.03 |
| WRDMEAc | 0.06 | 0.04 | 0.71 | 0.06 | -0.19 | 0.03 | -0.13 | 0.17 | -0.2 | 0.03 |
| WRDPOLc | 0.05 | 0.15 | 0.08 | 0.3 | -0.06 | 0.01 | -0.27 | 0.67 | 0.18 | 0.11 |
| WRDHYPn | 0.24 | 0.16 | 0.25 | 0.27 | -0.11 | -0.07 | 0.07 | 0.3 | 0.17 | -0.18 |
| WRDHYPv | 0.21 | 0 | 0.23 | 0.1 | -0.12 | 0.2 | -0.02 | 0.54 | 0.31 | -0.01 |
| WRDHYPnv | 0.13 | 0.08 | 0.61 | 0.11 | -0.23 | 0.22 | -0.34 | 0.12 | 0.24 | -0.05 |
| RDFRE | 0.82 | 0.19 | 0.04 | -0.11 | 0.05 | -0.19 | -0.09 | -0.1 | -0.32 | 0 |
| RDFKGL | -0.81 | -0.23 | -0.04 | 0.15 | -0.02 | 0.02 | 0.1 | 0.11 | 0.34 | 0.01 |
| RDL2 | 0.44 | 0.48 | -0.44 | 0.2 | -0.1 | -0.03 | -0.09 | 0.25 | 0.01 | -0.15 |
Coh-Metrix Model 1f
This model used principal component scores from 7 min narrative writing samples in spring (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * gbm = stochastic gradient boosted trees * pls = partial least squares regression * svm = support vector machines * enet = elastic net regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|
| -16.5845 | 0.1071 | 0.5091 | 0.6984 | -0.2708 | -0.0323 | 0.036 | -0.0221 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
PC1 = scores on 1st principal component extracted, …
Note: Importance is unavailable for support vector machines when PCA-based pre-processing is used (so all values for svm are 0).
| Metric | all | gbm | pls | svm | enet | rf | mars | cube |
|---|---|---|---|---|---|---|---|---|
| PC7 | 14.02 | 14.78 | 14.62 | 0 | 12.36 | 11.93 | 19.81 | 10 |
| PC1 | 13.63 | 30.49 | 12.95 | 0 | 6.54 | 26.18 | 26.11 | 20 |
| PC8 | 12.82 | 12.75 | 13.42 | 0 | 11.91 | 8.44 | 15.73 | 10 |
| PC6 | 10.25 | 12.69 | 10.89 | 0 | 7.93 | 8.88 | 10.09 | 16.6 |
| PC3 | 5.8 | 2.48 | 8 | 0 | 3.23 | 3.41 | 0.8 | 10 |
| PC9 | 4.73 | 3.43 | 5.34 | 0 | 4.67 | 2.71 | 0 | 6.6 |
| PC4 | 4.32 | 2.1 | 5.78 | 0 | 2.94 | 0.55 | 0.09 | 6.6 |
| PC16 | 3.92 | 1.27 | 3.57 | 0 | 6.22 | 0.83 | 0 | 3.4 |
| PC20 | 3.79 | 3.76 | 2.76 | 0 | 6.1 | 2.84 | 0 | 6.6 |
| PC24 | 3.35 | 2.04 | 2.26 | 0 | 6.11 | 5.13 | 0 | 3.4 |
| PC21 | 2.5 | 0 | 1.85 | 0 | 3.81 | 0.86 | 7.63 | 3.4 |
| PC32 | 2.2 | 0.74 | 1.2 | 0 | 5.05 | 1.77 | 0 | 0 |
| PC18 | 2.06 | 0.28 | 1.83 | 0 | 3.34 | 0.48 | 0 | 3.4 |
| PC15 | 2.06 | 0.86 | 2.12 | 0 | 2.84 | 1.45 | 0 | 0 |
| PC14 | 1.87 | 0.22 | 2.13 | 0 | 2.42 | 0.49 | 0 | 0 |
| PC19 | 1.76 | 0.83 | 1.58 | 0 | 2.82 | 1.1 | 0 | 0 |
| PC10 | 1.73 | 0.88 | 2.1 | 0 | 1.55 | 3.01 | 0 | 0 |
| PC31 | 1.69 | 0.48 | 1.03 | 0 | 3.81 | 0.38 | 0 | 0 |
| PC12 | 1.2 | 0.41 | 1.48 | 0 | 1.15 | 1.21 | 0 | 0 |
| PC30 | 1.06 | 0.58 | 0.76 | 0 | 1.97 | 1.51 | 0 | 0 |
| PC22 | 1.02 | 0.47 | 0.99 | 0 | 1.56 | 0.27 | 0 | 0 |
| PC17 | 0.83 | 1.24 | 0.22 | 0 | 0 | 3.14 | 13.07 | 0 |
| PC26 | 0.72 | 0.25 | 0.7 | 0 | 1.14 | 0.26 | 0 | 0 |
| PC5 | 0.58 | 1.09 | 0.47 | 0 | 0 | 2.15 | 4.42 | 0 |
| PC25 | 0.49 | 0.83 | 0.54 | 0 | 0.47 | 0.05 | 0 | 0 |
| PC13 | 0.46 | 0.3 | 0.67 | 0 | 0.07 | 2.34 | 0 | 0 |
| PC2 | 0.35 | 0.99 | 0.31 | 0 | 0 | 0 | 2.25 | 0 |
| PC11 | 0.23 | 1.78 | 0 | 0 | 0 | 3.42 | 0 | 0 |
| PC28 | 0.21 | 0.07 | 0.31 | 0 | 0 | 1.49 | 0 | 0 |
| PC29 | 0.11 | 0.55 | 0.08 | 0 | 0 | 0.75 | 0 | 0 |
| PC23 | 0.09 | 0.33 | 0.01 | 0 | 0 | 2.16 | 0 | 0 |
| PC33 | 0.07 | 0.57 | 0.03 | 0 | 0 | 0.32 | 0 | 0 |
| PC27 | 0.05 | 0.43 | 0 | 0 | 0 | 0.5 | 0 | 0 |
Proportion of Variance by Varimax Rotated Component (RC)
Due to space limitations, loadings for only the first ten principal components are displayed.
| Variable | RC1 | RC2 | RC4 | RC10 | RC5 | RC3 | RC8 | RC6 | RC7 | RC9 |
|---|---|---|---|---|---|---|---|---|---|---|
| SS loadings | 18.09 | 16.04 | 6.14 | 4.86 | 4.81 | 4.44 | 4.37 | 4.20 | 3.86 | 2.98 |
| Proportion Var | 0.19 | 0.17 | 0.06 | 0.05 | 0.05 | 0.05 | 0.05 | 0.04 | 0.04 | 0.03 |
| Cumulative Var | 0.19 | 0.35 | 0.42 | 0.47 | 0.51 | 0.56 | 0.61 | 0.65 | 0.69 | 0.72 |
| Proportion Explained | 0.26 | 0.23 | 0.09 | 0.07 | 0.07 | 0.06 | 0.06 | 0.06 | 0.06 | 0.04 |
| Cumulative Proportion | 0.26 | 0.49 | 0.58 | 0.65 | 0.72 | 0.78 | 0.84 | 0.90 | 0.96 | 1.00 |
Varimax Rotated Loadings
| Metric | RC1 | RC2 | RC4 | RC10 | RC5 | RC3 | RC8 | RC6 | RC7 | RC9 |
|---|---|---|---|---|---|---|---|---|---|---|
| DESSC | 0.85 | 0.02 | -0.07 | 0.2 | 0.07 | 0.05 | 0.04 | -0.04 | 0.13 | 0.11 |
| DESWC | 0.17 | 0.32 | 0.15 | 0.44 | -0.02 | 0.18 | 0.04 | -0.17 | 0.38 | 0.43 |
| DESPL | 0.85 | 0.02 | -0.07 | 0.2 | 0.07 | 0.05 | 0.04 | -0.04 | 0.13 | 0.11 |
| DESSL | -0.75 | -0.24 | 0.1 | 0.12 | -0.09 | 0.12 | -0.11 | -0.15 | 0.25 | 0.25 |
| DESSLd | 0.02 | 0.66 | 0.16 | 0.1 | -0.03 | 0.1 | 0.07 | 0.06 | -0.06 | 0.2 |
| DESWLsy | 0.09 | -0.08 | 0 | -0.05 | -0.14 | -0.16 | 0.86 | 0.02 | 0.06 | -0.13 |
| DESWLsyd | 0.03 | -0.07 | 0 | -0.04 | -0.07 | 0.06 | 0.82 | -0.1 | -0.04 | -0.09 |
| DESWLlt | 0.14 | -0.17 | -0.05 | -0.01 | -0.12 | -0.32 | 0.72 | 0.1 | 0.17 | -0.07 |
| DESWLltd | 0.12 | 0.06 | 0.09 | -0.05 | 0.15 | -0.05 | 0.74 | 0.06 | -0.13 | 0.18 |
| PCNARz | 0.35 | 0.7 | 0.16 | 0.21 | 0.45 | 0.21 | -0.14 | 0.03 | -0.03 | 0.17 |
| PCNARp | 0.47 | 0.57 | 0.1 | 0.27 | 0.41 | 0.2 | -0.13 | -0.02 | -0.02 | 0.16 |
| PCSYNz | 0.89 | -0.07 | -0.03 | -0.17 | 0.08 | -0.17 | 0.09 | 0.06 | -0.07 | -0.23 |
| PCSYNp | 0.88 | -0.13 | 0.05 | -0.15 | 0.01 | -0.14 | 0.02 | 0.04 | 0.08 | -0.11 |
| PCCNCz | -0.64 | -0.38 | -0.18 | -0.4 | -0.06 | -0.31 | -0.04 | -0.22 | 0.07 | 0.23 |
| PCCNCp | -0.48 | -0.13 | -0.1 | -0.45 | -0.04 | -0.36 | 0.03 | -0.14 | 0.05 | 0.22 |
| PCREFz | -0.4 | 0.72 | 0.01 | -0.14 | 0.29 | 0.09 | -0.24 | -0.13 | 0.1 | -0.1 |
| PCREFp | -0.49 | 0.4 | -0.04 | -0.04 | 0.4 | 0.15 | -0.24 | -0.1 | 0.05 | -0.05 |
| PCDCz | 0.17 | 0.3 | 0.9 | 0.06 | 0.06 | 0.07 | 0 | 0.06 | 0.13 | 0.02 |
| PCDCp | 0.18 | 0.32 | 0.79 | 0.15 | 0.09 | 0.08 | 0.05 | 0.04 | 0.09 | 0.07 |
| PCVERBz | -0.62 | -0.62 | -0.01 | -0.1 | -0.18 | 0.3 | -0.09 | -0.05 | 0.2 | -0.09 |
| PCVERBp | -0.41 | -0.49 | 0.06 | -0.17 | -0.22 | 0.43 | -0.1 | -0.03 | 0.16 | -0.11 |
| PCCONNz | 0.24 | 0.02 | 0.09 | -0.06 | -0.06 | 0.08 | -0.01 | 0.9 | 0.11 | 0.03 |
| PCCONNp | 0.13 | -0.07 | 0.03 | -0.24 | 0.12 | -0.05 | -0.12 | 0.75 | 0.11 | -0.14 |
| PCTEMPz | 0.72 | 0.62 | 0.09 | 0.09 | 0.01 | 0.01 | 0.13 | 0.05 | 0.05 | 0.12 |
| PCTEMPp | 0.47 | 0.54 | 0.14 | 0.17 | -0.11 | 0.02 | 0.17 | 0.07 | 0 | 0.17 |
| CRFNO1 | 0.07 | 0.7 | 0.15 | 0.07 | -0.37 | -0.13 | -0.22 | -0.03 | 0.11 | -0.07 |
| CRFAO1 | 0.36 | 0.85 | 0.06 | 0.07 | 0.17 | 0.02 | 0.11 | 0 | 0.06 | 0.06 |
| CRFSO1 | 0.09 | 0.77 | 0.1 | -0.01 | -0.32 | -0.16 | -0.22 | -0.07 | 0.06 | -0.11 |
| CRFNOa | -0.03 | 0.77 | 0.08 | 0.01 | -0.32 | -0.08 | -0.21 | -0.05 | 0.08 | -0.1 |
| CRFAOa | 0.3 | 0.86 | 0.07 | 0.08 | 0.25 | 0.07 | 0.12 | 0.02 | 0.05 | 0.06 |
| CRFSOa | -0.01 | 0.81 | 0.07 | -0.04 | -0.28 | -0.12 | -0.2 | -0.09 | 0.05 | -0.1 |
| CRFCWO1 | 0.27 | 0.89 | 0.08 | -0.08 | 0.17 | 0.05 | -0.01 | -0.04 | 0.08 | -0.03 |
| CRFCWO1d | 0.81 | 0.27 | -0.05 | 0.07 | -0.03 | 0.02 | 0.02 | -0.04 | 0.03 | 0.06 |
| CRFCWOa | 0.18 | 0.9 | 0.08 | -0.07 | 0.19 | 0.07 | 0 | -0.04 | 0.07 | -0.04 |
| CRFCWOad | 0.83 | 0.33 | -0.02 | 0.12 | -0.01 | 0.02 | 0.02 | 0.02 | 0.03 | 0.1 |
| CRFANP1 | 0.38 | 0.8 | 0.09 | 0.08 | 0.29 | 0.05 | 0.12 | 0.03 | 0.03 | 0.08 |
| CRFANPa | 0.18 | 0.84 | 0.11 | 0.04 | 0.29 | 0.11 | 0.15 | 0.05 | 0.02 | 0.04 |
| LSASS1 | 0.29 | 0.81 | 0.07 | -0.08 | 0.11 | -0.13 | -0.14 | -0.03 | 0.03 | 0.01 |
| LSASS1d | 0.67 | 0.34 | 0.01 | 0.13 | -0.09 | -0.13 | -0.16 | 0.02 | 0.03 | 0.1 |
| LSASSp | 0.19 | 0.85 | 0.06 | -0.1 | 0.11 | -0.07 | -0.09 | -0.03 | 0.03 | -0.02 |
| LSASSpd | 0.76 | 0.38 | -0.02 | 0.09 | -0.12 | -0.1 | -0.1 | 0.06 | 0.05 | 0.09 |
| LSAGN | 0.73 | 0.58 | -0.01 | 0.13 | 0.09 | -0.05 | -0.01 | -0.04 | 0.12 | 0.04 |
| LSAGNd | 0.59 | 0.72 | 0.03 | 0.02 | -0.05 | -0.08 | -0.07 | 0 | 0.04 | 0.02 |
| LDTTRc | -0.11 | -0.48 | 0.03 | -0.19 | 0.15 | -0.09 | 0.22 | 0.27 | -0.56 | 0.16 |
| LDTTRa | -0.03 | -0.46 | 0.01 | -0.15 | -0.02 | -0.31 | 0.18 | 0.5 | -0.42 | -0.11 |
| LDMTLD | 0.19 | -0.13 | 0.15 | 0.25 | -0.03 | -0.16 | 0.28 | 0.37 | -0.19 | 0.32 |
| CNCAll | -0.3 | 0.07 | 0.62 | -0.12 | 0.17 | -0.01 | -0.06 | -0.62 | -0.02 | -0.09 |
| CNCCaus | 0.04 | 0.02 | 0.86 | -0.19 | 0 | 0.12 | 0.13 | 0.03 | -0.01 | -0.08 |
| CNCLogic | -0.09 | 0.11 | 0.78 | 0.31 | 0.08 | 0.06 | -0.09 | -0.01 | -0.05 | -0.13 |
| CNCADC | 0.05 | -0.11 | -0.1 | 0.71 | -0.04 | 0.06 | 0.12 | -0.24 | -0.16 | 0.05 |
| CNCTemp | 0.05 | 0.24 | 0.35 | 0.16 | 0.09 | -0.15 | -0.26 | 0.14 | 0.14 | 0.03 |
| CNCTempx | -0.02 | 0.11 | 0.14 | 0.09 | -0.16 | 0.33 | 0.08 | -0.01 | -0.19 | 0.25 |
| CNCAdd | -0.36 | -0.05 | 0 | -0.12 | 0.11 | -0.07 | -0.09 | -0.84 | -0.11 | -0.05 |
| CNCPos | -0.27 | 0.11 | 0.61 | -0.31 | 0.19 | -0.05 | -0.1 | -0.53 | 0.03 | -0.1 |
| CNCNeg | 0 | -0.14 | -0.08 | 0.63 | -0.06 | 0.08 | 0.07 | -0.22 | -0.17 | 0.02 |
| SMCAUSv | 0.76 | -0.13 | 0.04 | -0.2 | 0.05 | -0.1 | 0.23 | 0.11 | 0.13 | 0.06 |
| SMCAUSvp | 0.58 | -0.1 | 0.55 | -0.22 | 0.01 | -0.03 | 0.22 | 0.15 | 0.13 | -0.03 |
| SMINTEp | 0.62 | 0.03 | -0.13 | -0.29 | 0.33 | -0.08 | -0.06 | 0.16 | 0.08 | -0.01 |
| SMCAUSr | -0.25 | 0.24 | 0.61 | 0.15 | -0.05 | 0.2 | -0.11 | 0.04 | 0.1 | 0.13 |
| SMINTEr | -0.23 | 0.03 | 0.66 | 0.03 | -0.13 | 0.26 | 0.11 | -0.11 | 0.13 | 0.18 |
| SMCAUSlsa | -0.16 | 0.03 | -0.08 | -0.11 | -0.21 | 0.43 | 0.17 | -0.07 | 0.38 | -0.38 |
| SMCAUSwn | 0.11 | 0.06 | 0.17 | -0.08 | 0.17 | 0.12 | 0.02 | 0.09 | 0.71 | 0.06 |
| SMTEMP | 0.73 | 0.62 | 0.07 | 0.08 | 0 | 0.01 | 0.13 | 0.05 | 0.05 | 0.12 |
| SYNLE | -0.17 | -0.03 | -0.01 | 0.03 | 0.02 | -0.18 | 0.08 | 0.23 | 0.35 | 0.02 |
| SYNNP | -0.03 | -0.11 | -0.04 | -0.14 | -0.65 | -0.04 | 0.11 | 0.03 | 0 | 0.01 |
| SYNMEDpos | 0.74 | 0.57 | 0.07 | 0.11 | 0.01 | 0.02 | 0.12 | 0.04 | 0.04 | 0.12 |
| SYNMEDwrd | 0.76 | 0.56 | 0.07 | 0.11 | 0 | 0 | 0.14 | 0.07 | 0.05 | 0.13 |
| SYNMEDlem | 0.76 | 0.55 | 0.07 | 0.11 | -0.01 | 0 | 0.14 | 0.08 | 0.05 | 0.14 |
| SYNSTRUTa | 0.77 | 0.18 | -0.04 | -0.11 | 0.04 | 0.05 | 0.05 | 0.15 | 0.07 | -0.01 |
| SYNSTRUTt | 0.82 | 0.16 | -0.08 | -0.11 | 0.04 | 0.04 | 0.01 | 0.18 | 0.08 | 0.01 |
| DRNP | -0.17 | 0.01 | -0.27 | -0.2 | -0.15 | 0.04 | -0.25 | 0.33 | -0.36 | 0.1 |
| DRVP | 0.16 | 0.04 | 0.09 | -0.05 | 0.64 | -0.04 | 0.03 | -0.04 | 0.22 | -0.22 |
| DRAP | -0.1 | 0.09 | 0.25 | 0.55 | 0.08 | -0.01 | -0.11 | -0.01 | 0.25 | -0.07 |
| DRPP | 0.04 | -0.06 | -0.09 | -0.13 | -0.13 | 0 | 0.02 | 0.18 | 0 | 0.66 |
| DRNEG | 0.03 | 0.03 | -0.11 | 0.5 | 0.05 | 0.06 | -0.08 | 0.13 | -0.12 | -0.06 |
| WRDNOUN | 0.05 | -0.06 | -0.13 | -0.26 | -0.71 | -0.31 | 0.03 | 0.24 | 0.05 | 0.02 |
| WRDVERB | 0.23 | 0.1 | 0.08 | -0.06 | 0.61 | -0.04 | 0.07 | 0.07 | 0.21 | 0.07 |
| WRDADJ | 0.09 | -0.11 | -0.09 | 0.09 | -0.24 | 0.1 | 0.31 | 0.1 | -0.1 | -0.56 |
| WRDADV | -0.09 | 0.14 | 0.33 | 0.71 | 0.06 | 0.06 | -0.1 | -0.05 | 0.19 | -0.08 |
| WRDPRO | -0.04 | 0.14 | -0.08 | 0 | 0.61 | 0.27 | -0.34 | 0.06 | -0.27 | 0.02 |
| WRDPRP3s | 0.03 | 0.12 | 0 | 0.21 | 0.06 | -0.03 | -0.04 | 0.08 | 0.01 | 0.09 |
| WRDPRP3p | -0.06 | -0.18 | -0.05 | -0.03 | 0 | -0.17 | 0.07 | 0.18 | 0.04 | -0.3 |
| WRDFRQc | -0.08 | -0.06 | 0.23 | 0.22 | 0.16 | 0.75 | -0.34 | 0.07 | 0.04 | -0.14 |
| WRDFRQa | -0.26 | -0.09 | 0.17 | 0.07 | 0.13 | 0.58 | -0.43 | -0.17 | -0.25 | 0.22 |
| WRDFRQmc | 0.79 | 0.09 | 0.01 | 0.04 | 0.12 | 0.07 | 0.05 | 0 | 0.09 | 0.08 |
| WRDAOAc | 0.22 | -0.06 | 0.11 | 0.24 | -0.11 | -0.2 | 0.07 | 0.1 | 0.07 | -0.14 |
| WRDFAMc | -0.02 | -0.03 | 0.12 | 0.02 | 0.12 | 0.67 | -0.08 | 0.09 | 0.19 | 0.09 |
| WRDCNCc | 0.04 | 0.03 | -0.31 | -0.41 | -0.07 | -0.5 | 0.03 | 0.02 | 0.1 | 0.51 |
| WRDIMGc | 0.01 | -0.02 | -0.38 | -0.48 | -0.16 | -0.48 | 0.12 | -0.01 | 0.05 | 0.38 |
| WRDMEAc | 0.11 | -0.11 | -0.36 | -0.47 | -0.13 | -0.18 | 0.25 | -0.1 | 0.09 | 0.37 |
| WRDPOLc | 0.15 | 0.08 | 0.25 | -0.21 | 0.13 | 0.2 | -0.04 | 0.15 | 0.67 | -0.04 |
| WRDHYPn | 0.11 | 0.25 | 0.03 | 0.04 | 0.22 | -0.32 | -0.03 | 0.07 | 0.48 | 0.07 |
| WRDHYPv | 0.14 | 0.04 | -0.01 | -0.07 | 0.57 | -0.12 | 0.01 | -0.03 | 0.46 | 0.06 |
| WRDHYPnv | 0.04 | 0.16 | -0.1 | -0.23 | -0.23 | -0.52 | -0.02 | 0.14 | 0.49 | 0.03 |
| RDFRE | 0.79 | 0.26 | -0.08 | -0.09 | 0.14 | -0.09 | -0.13 | 0.11 | -0.23 | -0.18 |
| RDFKGL | -0.75 | -0.25 | 0.1 | 0.11 | -0.11 | 0.11 | -0.03 | -0.15 | 0.26 | 0.24 |
| RDL2 | 0.48 | 0.59 | 0.15 | 0.01 | 0.2 | 0.44 | -0.16 | 0.08 | 0.1 | -0.1 |
Coh-Metrix Model 2
General Description
Coh-Metrix Model 2 is a simplified version of Model 1. Model 2 is recommended for use over Model 1.
Coh-Metrix Model 2 is an ensemble (formed by averaging predicted quality scores) of the three sub-models described below.
Highly correlated Coh-Metrix metrics (r > |.90|) were excluded during pre-processing (see section on Scoring Model Development for more details).
All of these models used Coh-Metrix scores on 7 min narrative writing samples (“I once had a magic pencil and …”) from students in the fall, winter, and spring of Grades 2-5 (Mercer et al., 2019) to predict holistic writing quality on the samples (elo ratings calculated from paired comparisons). More details on the sample are available in (Mercer et al., 2019).
Coh-Metrix Model 2a
This model was trained on fall data in (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * pls = partial least squares regression * rf = random forest regression * mars = bagged multivariate adaptive regression splines * gbm = stochastic gradient boosted trees * svm = support vector machines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | rf | mars | gbm | svm | cube |
|---|---|---|---|---|---|---|
| -11.0081 | 0.1741 | 0.0413 | 0.1875 | 0.2353 | 0.206 | 0.2108 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | rf | mars | gbm | svm | cube |
|---|---|---|---|---|---|---|---|
| DESWC | 29.91 | 5.86 | 14.51 | 55.78 | 44.87 | 5.96 | 36.49 |
| DESWLlt | 8.67 | 2.98 | 2.69 | 18.79 | 3.07 | 2.39 | 17.89 |
| LDMTLD | 7.35 | 4.01 | 5.57 | 0 | 9.55 | 3.9 | 17.89 |
| WRDHYPn | 7.16 | 2.9 | 2.68 | 9.55 | 3.85 | 2.29 | 17.89 |
| LDTTRa | 2.84 | 2.01 | 0.97 | 11.14 | 0.26 | 1.12 | 1.05 |
| CNCPos | 1.36 | 0.88 | 1.11 | 4.75 | 0.05 | 1.28 | 0.35 |
| DESWLsy | 1.3 | 2.09 | 1.9 | 0 | 1.66 | 1.54 | 1.05 |
| CNCTempx | 1.11 | 0.89 | 1.03 | 0 | 1.63 | 2.48 | 0.35 |
| CNCLogic | 1.09 | 1.42 | 1.39 | 0 | 1.81 | 1.7 | 0.35 |
| PCDCp | 1.08 | 2 | 2.56 | 0 | 1.71 | 1.36 | 0 |
| DESPL | 1.04 | 3.22 | 2.18 | 0 | 0.04 | 2.15 | 0 |
| DESWLltd | 1.02 | 2.3 | 1.13 | 0 | 1.26 | 1.6 | 0 |
| WRDFRQa | 0.96 | 1.86 | 0.58 | 0 | 0.17 | 1.97 | 1.05 |
| DESWLsyd | 0.92 | 1.97 | 1.87 | 0 | 1.21 | 1.29 | 0 |
| DESSLd | 0.9 | 1.67 | 2 | 0 | 1.27 | 1.36 | 0 |
| CNCTemp | 0.89 | 0.93 | 1.39 | 0 | 1.08 | 1.89 | 0.35 |
| LSAGN | 0.87 | 2.55 | 1.18 | 0 | 0.22 | 1.8 | 0 |
| LSASSpd | 0.86 | 1.95 | 0.86 | 0 | 0.36 | 1.79 | 0.35 |
| CNCADC | 0.85 | 1.12 | 1.41 | 0 | 0.67 | 2.37 | 0 |
| DRPP | 0.84 | 2.18 | 2.11 | 0 | 0.57 | 1.4 | 0 |
| PCCONNz | 0.82 | 1.09 | 0.4 | 0 | 1.61 | 1.36 | 0 |
| WRDPRO | 0.82 | 1.77 | 1.44 | 0 | 0.73 | 1.61 | 0 |
| SYNSTRUTa | 0.8 | 0.85 | 3.07 | 0 | 0.58 | 1.4 | 0.7 |
| CRFCWO1d | 0.79 | 1.67 | 1.39 | 0 | 0.43 | 1.88 | 0 |
| LSASS1d | 0.78 | 1.55 | 0.48 | 0 | 0.78 | 1.72 | 0 |
| SMCAUSwn | 0.76 | 1.4 | 1.04 | 0 | 0.31 | 2.16 | 0 |
| SYNMEDpos | 0.75 | 1.75 | 0.6 | 0 | 0.77 | 1.36 | 0 |
| SMINTEp | 0.73 | 0.75 | 0.85 | 0 | 0.79 | 1.66 | 0.35 |
| LDTTRc | 0.73 | 1.82 | 0.7 | 0 | 0.72 | 1.23 | 0 |
| CRFCWOad | 0.73 | 1.51 | 1.12 | 0 | 0.38 | 1.81 | 0 |
| WRDVERB | 0.71 | 1.04 | 0.93 | 0 | 0.58 | 0.86 | 1.05 |
| WRDFAMc | 0.7 | 1.09 | 0.5 | 0 | 1.32 | 1.08 | 0 |
| WRDHYPnv | 0.69 | 1.9 | 0.91 | 0 | 0.13 | 1.22 | 0.35 |
| WRDFRQmc | 0.69 | 1.35 | 2.5 | 0 | 1.3 | 0.4 | 0 |
| WRDCNCc | 0.69 | 1.07 | 0.44 | 0 | 0.58 | 0.8 | 1.05 |
| PCNARz | 0.68 | 1.29 | 1.29 | 0 | 0.58 | 1.1 | 0.35 |
| WRDPOLc | 0.67 | 0.85 | 0.77 | 0 | 0.51 | 1.97 | 0 |
| RDFRE | 0.66 | 1.29 | 0.91 | 0 | 0.76 | 1.23 | 0 |
| CRFNOa | 0.63 | 0.74 | 1.34 | 0 | 0.56 | 1.67 | 0 |
| PCVERBz | 0.63 | 1.55 | 0.67 | 0 | 0.28 | 1.09 | 0.35 |
| LSAGNd | 0.61 | 1.5 | 0.95 | 0 | 0.24 | 1.38 | 0 |
| SMCAUSvp | 0.59 | 0.93 | 0.73 | 0 | 0.36 | 1.67 | 0 |
| WRDADV | 0.58 | 1.3 | 0.95 | 0 | 0.13 | 1.55 | 0 |
| WRDAOAc | 0.58 | 1.66 | 0.46 | 0 | 0.27 | 1.18 | 0 |
| DRNP | 0.57 | 1.73 | 1.5 | 0 | 0.27 | 0.82 | 0 |
| WRDHYPv | 0.56 | 0.28 | 1.01 | 0 | 0.83 | 1.47 | 0 |
| DRVP | 0.56 | 0.82 | 0.62 | 0 | 0.82 | 0.75 | 0.35 |
| PCNARp | 0.54 | 1.76 | 1.29 | 0 | 0 | 1 | 0 |
| CNCCaus | 0.51 | 1.11 | 0.58 | 0 | 0.11 | 1.46 | 0 |
| CRFCWOa | 0.49 | 0.46 | 0.56 | 0 | 0.37 | 1.57 | 0 |
| SMCAUSr | 0.47 | 1.58 | 1.07 | 0 | 0.47 | 0.29 | 0 |
| SMCAUSlsa | 0.46 | 0.24 | 0.86 | 0 | 0.63 | 1.26 | 0 |
| LSASSp | 0.46 | 0.79 | 1.01 | 0 | 0.15 | 1.31 | 0 |
| SMINTEr | 0.45 | 1.83 | 0.9 | 0 | 0.13 | 0.45 | 0 |
| DRAP | 0.44 | 0.46 | 1.01 | 0 | 0.41 | 1.17 | 0 |
| DRNEG | 0.44 | 0.76 | 0.43 | 0 | 0.09 | 1.41 | 0 |
| WRDNOUN | 0.43 | 0.56 | 0.96 | 0 | 0.75 | 0.66 | 0 |
| WRDFRQc | 0.43 | 0.54 | 0.67 | 0 | 0.48 | 1.04 | 0 |
| WRDPRP3s | 0.43 | 0.76 | 0.84 | 0 | 0.74 | 0.56 | 0 |
| CRFANPa | 0.41 | 0.76 | 1.36 | 0 | 0.18 | 0.96 | 0 |
| SYNLE | 0.4 | 1.13 | 0.95 | 0 | 0.1 | 0.77 | 0 |
| PCTEMPp | 0.4 | 1.05 | 0.22 | 0 | 0.55 | 0.47 | 0 |
| WRDADJ | 0.39 | 0.99 | 0.94 | 0 | 0.2 | 0.74 | 0 |
| RDL2 | 0.39 | 0.31 | 1.06 | 0 | 0.39 | 1.09 | 0 |
| WRDIMGc | 0.37 | 0.14 | 0.74 | 0 | 0.3 | 0.93 | 0.35 |
| PCVERBp | 0.36 | 0.99 | 1.39 | 0 | 0 | 0.75 | 0 |
| PCCNCz | 0.35 | 1.18 | 0.1 | 0 | 0.32 | 0.39 | 0 |
| WRDMEAc | 0.29 | 0.21 | 0.45 | 0 | 0.44 | 0.71 | 0 |
| PCREFz | 0.29 | 0.47 | 1.07 | 0 | 0.3 | 0.53 | 0 |
| SMCAUSv | 0.25 | 0.39 | 0.77 | 0 | 0.13 | 0.66 | 0 |
| SYNNP | 0.22 | 0.03 | 0.64 | 0 | 0.58 | 0.33 | 0 |
| PCSYNp | 0.21 | 0.68 | 1.55 | 0 | 0.15 | 0 | 0 |
| PCCONNp | 0.17 | 0 | 0.33 | 0 | 0.11 | 0.66 | 0 |
| PCCNCp | 0.16 | 0.57 | 0.78 | 0 | 0 | 0.16 | 0 |
| PCREFp | 0.15 | 0.06 | 0.74 | 0 | 0 | 0.58 | 0 |
| WRDPRP3p | 0.14 | 0.84 | 0 | 0 | 0 | 0.03 | 0 |
Coh-Metrix Model 2b
This model was trained on winter data (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * mars = bagged multivariate adaptive regression splines * gbm = stochastic gradient boosted trees * svm = support vector machines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | mars | gbm | svm | cube |
|---|---|---|---|---|
| -7.2585 | 0.2289 | 0.5300 | 0.1527 | 0.1150 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | mars | gbm | svm | cube |
|---|---|---|---|---|---|
| DESWC | 30.39 | 45.46 | 34.5 | 4.37 | 16.04 |
| LSAGN | 7.18 | 0 | 9.31 | 2.85 | 17.43 |
| DESWLlt | 6.73 | 19 | 2.42 | 1.37 | 9.31 |
| LDMTLD | 5.59 | 0 | 8.91 | 2.47 | 5.54 |
| SYNLE | 5.43 | 13.58 | 3.65 | 1.79 | 2.18 |
| WRDIMGc | 3.6 | 9.36 | 1.84 | 1.22 | 3.37 |
| WRDNOUN | 3.19 | 6.64 | 1.38 | 1.77 | 6.53 |
| CNCAdd | 1.87 | 5.96 | 0.45 | 1.02 | 1.39 |
| WRDVERB | 1.42 | 0 | 2.02 | 1.67 | 1.19 |
| SMCAUSwn | 1.25 | 0 | 1.83 | 2.04 | 0 |
| DESWLltd | 1.16 | 0 | 1.21 | 1.53 | 2.77 |
| CRFCWO1d | 1.09 | 0 | 1.18 | 2.17 | 1.39 |
| WRDHYPnv | 1.02 | 0 | 0.98 | 1.36 | 2.77 |
| CRFNOa | 0.99 | 0 | 1.37 | 1.9 | 0 |
| RDFRE | 0.99 | 0 | 1.54 | 0.25 | 1.39 |
| LSAGNd | 0.85 | 0 | 0.53 | 1.76 | 2.77 |
| DESWLsy | 0.77 | 0 | 1.11 | 1.28 | 0 |
| SYNMEDpos | 0.76 | 0 | 0.29 | 2 | 2.77 |
| WRDPRP3s | 0.74 | 0 | 0.91 | 1.53 | 0.4 |
| DESPL | 0.73 | 0 | 0.44 | 2.34 | 1.39 |
| CNCAll | 0.72 | 0 | 0.44 | 0.96 | 3.17 |
| RDL2 | 0.71 | 0 | 0.6 | 1.65 | 1.39 |
| PCCNCz | 0.71 | 0 | 0.62 | 1.6 | 1.39 |
| PCVERBz | 0.69 | 0 | 0.82 | 1.78 | 0 |
| WRDPRO | 0.69 | 0 | 0.69 | 1.18 | 1.39 |
| CNCTemp | 0.65 | 0 | 0.71 | 1.87 | 0 |
| WRDFRQc | 0.65 | 0 | 0.98 | 0.99 | 0 |
| WRDFRQmc | 0.63 | 0 | 0.56 | 1.25 | 1.39 |
| DRVP | 0.62 | 0 | 0.93 | 0.92 | 0 |
| LSASS1d | 0.61 | 0 | 0.65 | 1.87 | 0 |
| SMCAUSlsa | 0.6 | 0 | 0.94 | 0.77 | 0 |
| CRFCWOad | 0.6 | 0 | 0.61 | 1.92 | 0 |
| WRDMEAc | 0.59 | 0 | 0.53 | 1.4 | 0.99 |
| PCTEMPp | 0.56 | 0 | 0.78 | 1.02 | 0 |
| SMCAUSv | 0.56 | 0 | 0.85 | 0.83 | 0 |
| PCCNCp | 0.56 | 0 | 0.02 | 1.62 | 2.77 |
| WRDFRQa | 0.53 | 0 | 0.73 | 1.01 | 0 |
| LSASSp | 0.53 | 0 | 0.54 | 1.71 | 0 |
| LDTTRc | 0.52 | 0 | 0.64 | 1.3 | 0 |
| DRPP | 0.51 | 0 | 0.7 | 1.02 | 0 |
| PCREFp | 0.5 | 0 | 0 | 0.7 | 3.56 |
| CRFCWO1 | 0.47 | 0 | 0.4 | 1.75 | 0 |
| SMCAUSvp | 0.46 | 0 | 0.47 | 1.42 | 0 |
| PCNARz | 0.45 | 0 | 0.26 | 1.64 | 0.59 |
| SYNNP | 0.45 | 0 | 0.3 | 0.91 | 1.39 |
| PCSYNz | 0.45 | 0 | 0.32 | 0.87 | 1.39 |
| SMINTEp | 0.45 | 0 | 0.4 | 1.66 | 0 |
| LDTTRa | 0.43 | 0 | 0.23 | 1.06 | 1.39 |
| DESWLsyd | 0.42 | 0 | 0.35 | 1.62 | 0 |
| CRFANPa | 0.41 | 0 | 0.34 | 1.59 | 0 |
| SMINTEr | 0.41 | 0 | 0.6 | 0.69 | 0 |
| CNCLogic | 0.4 | 0 | 0.51 | 0.89 | 0 |
| WRDAOAc | 0.4 | 0 | 0.58 | 0.69 | 0 |
| WRDHYPv | 0.4 | 0 | 0.36 | 1.45 | 0 |
| CNCNeg | 0.4 | 0 | 0.32 | 1.17 | 0.59 |
| CNCCaus | 0.38 | 0 | 0.47 | 0.92 | 0 |
| WRDFAMc | 0.37 | 0 | 0.46 | 0.91 | 0 |
| SYNSTRUTa | 0.36 | 0 | 0.36 | 1.2 | 0 |
| CRFAOa | 0.35 | 0 | 0.19 | 1.68 | 0 |
| WRDADV | 0.34 | 0 | 0.32 | 1.19 | 0 |
| SMCAUSr | 0.33 | 0 | 0.61 | 0.1 | 0 |
| DESSLd | 0.32 | 0 | 0.18 | 1.5 | 0 |
| PCCONNp | 0.29 | 0 | 0.46 | 0.37 | 0 |
| WRDPOLc | 0.28 | 0 | 0.28 | 0.93 | 0 |
| WRDADJ | 0.28 | 0 | 0.4 | 0.51 | 0 |
| DRAP | 0.26 | 0 | 0.24 | 0.87 | 0 |
| DRNP | 0.26 | 0 | 0.33 | 0.57 | 0 |
| WRDHYPn | 0.26 | 0 | 0.27 | 0.84 | 0 |
| CNCTempx | 0.25 | 0 | 0.23 | 0.86 | 0 |
| DRNEG | 0.23 | 0 | 0.13 | 1.08 | 0 |
| PCREFz | 0.23 | 0 | 0.24 | 0.7 | 0 |
| PCVERBp | 0.22 | 0 | 0 | 1.48 | 0 |
| PCNARp | 0.2 | 0 | 0.01 | 1.35 | 0 |
| PCDCp | 0.19 | 0 | 0.11 | 0.9 | 0 |
| PCSYNp | 0.09 | 0 | 0.02 | 0.56 | 0 |
| WRDPRP3p | 0 | 0 | 0 | 0 | 0 |
Coh-Metrix Model 2c
This model was trained on spring data (Mercer et al., 2019).
Algorithm Weightings in Ensemble
Abbreviations: * all = ensemble model * pls = partial least squares regression * mars = bagged multivariate adaptive regression splines * gbm = stochastic gradient boosted trees * svm = support vector machines
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | mars | gbm | svm |
|---|---|---|---|---|
| -10.8192 | 0.0374 | 0.2735 | 0.243 | 0.5377 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | mars | gbm | svm |
|---|---|---|---|---|---|
| DESWC | 22.58 | 5.76 | 48.45 | 37.37 | 3.91 |
| WRDVERB | 7.27 | 1.88 | 23.22 | 2.92 | 1.5 |
| DESWLltd | 5.72 | 1.75 | 16.17 | 3.85 | 1.53 |
| CRFCWOa | 4.57 | 1.9 | 12.16 | 1.14 | 2.44 |
| PCNARp | 2.04 | 2.72 | 0 | 3.23 | 2.49 |
| PCDCz | 1.78 | 1.05 | 0 | 3.24 | 2.07 |
| CRFANPa | 1.67 | 1.23 | 0 | 2.21 | 2.3 |
| LSASS1d | 1.67 | 1.5 | 0 | 1.62 | 2.56 |
| WRDHYPn | 1.59 | 2.38 | 0 | 3.29 | 1.58 |
| SYNSTRUTa | 1.59 | 1.74 | 0 | 1.28 | 2.54 |
| LDMTLD | 1.57 | 1.79 | 0 | 2.47 | 1.95 |
| LSAGN | 1.57 | 1.72 | 0 | 1.17 | 2.55 |
| SMCAUSvp | 1.53 | 0.97 | 0 | 1.91 | 2.17 |
| DESSLd | 1.52 | 0.87 | 0 | 1.28 | 2.45 |
| LSAGNd | 1.49 | 2.29 | 0 | 0.11 | 2.81 |
| WRDFRQmc | 1.41 | 2.04 | 0 | 1.45 | 2.06 |
| DESPL | 1.41 | 3.29 | 0 | 0.85 | 2.25 |
| PCVERBz | 1.25 | 1.73 | 0 | 0.62 | 2.13 |
| SYNMEDpos | 1.24 | 1.78 | 0 | 0.72 | 2.08 |
| LSASSp | 1.22 | 1.97 | 0 | 0.13 | 2.28 |
| SMCAUSv | 1.16 | 0.91 | 0 | 1.16 | 1.78 |
| CRFCWO1d | 1.15 | 1.54 | 0 | 0.21 | 2.14 |
| SMCAUSwn | 1.09 | 2.26 | 0 | 0.94 | 1.63 |
| CNCTempx | 1.08 | 0.26 | 0 | 0.57 | 1.92 |
| WRDHYPv | 1.06 | 2.37 | 0 | 1.41 | 1.36 |
| SMCAUSlsa | 1.03 | 1.59 | 0 | 1.07 | 1.5 |
| WRDNOUN | 1 | 2.13 | 0 | 1.68 | 1.11 |
| PCDCp | 1 | 1.81 | 0 | 0.23 | 1.8 |
| PCVERBp | 0.93 | 1.09 | 0 | 0.05 | 1.79 |
| PCTEMPp | 0.92 | 1.68 | 0 | 0.51 | 1.52 |
| LDTTRc | 0.9 | 1.26 | 0 | 1.34 | 1.14 |
| WRDPRP3s | 0.87 | 1.36 | 0 | 1.67 | 0.92 |
| DESWLlt | 0.85 | 2.05 | 0 | 1.12 | 1.07 |
| CNCTemp | 0.85 | 0.85 | 0 | 0.9 | 1.27 |
| RDL2 | 0.84 | 2.17 | 0 | 0.54 | 1.31 |
| DRPP | 0.83 | 1.78 | 0 | 1.37 | 0.94 |
| PCCNCz | 0.8 | 1.87 | 0 | 0.31 | 1.35 |
| DRNP | 0.8 | 1.62 | 0 | 0.79 | 1.16 |
| LDTTRa | 0.79 | 2.31 | 0 | 0.24 | 1.34 |
| WRDAOAc | 0.78 | 0.69 | 0 | 0.78 | 1.17 |
| RDFKGL | 0.73 | 2.06 | 0 | 0.13 | 1.27 |
| SYNNP | 0.68 | 0.65 | 0 | 0.24 | 1.23 |
| CNCPos | 0.68 | 0.82 | 0 | 0.67 | 1.03 |
| CNCCaus | 0.67 | 0.73 | 0 | 0.63 | 1.02 |
| WRDADV | 0.67 | 1.9 | 0 | 0.66 | 0.93 |
| PCSYNz | 0.66 | 1.76 | 0 | 0.33 | 1.07 |
| WRDPRO | 0.64 | 0.73 | 0 | 0.9 | 0.84 |
| CNCLogic | 0.64 | 0.41 | 0 | 0.71 | 0.95 |
| PCCNCp | 0.64 | 1.15 | 0 | 0 | 1.22 |
| DRVP | 0.63 | 0.79 | 0 | 0.32 | 1.08 |
| WRDADJ | 0.62 | 1.12 | 0 | 0.38 | 1 |
| SMINTEp | 0.62 | 1.06 | 0 | 0.53 | 0.95 |
| DRNEG | 0.6 | 0.73 | 0 | 0.06 | 1.13 |
| WRDPOLc | 0.6 | 0.89 | 0 | 0.61 | 0.88 |
| WRDHYPnv | 0.58 | 0.02 | 0 | 0.17 | 1.09 |
| WRDFRQa | 0.58 | 0.36 | 0 | 0.35 | 0.99 |
| SYNLE | 0.57 | 0.09 | 0 | 0.62 | 0.87 |
| SMCAUSr | 0.56 | 0.05 | 0 | 0.3 | 1 |
| DRAP | 0.55 | 1.23 | 0 | 0.32 | 0.89 |
| PCREFz | 0.53 | 1.2 | 0 | 0.46 | 0.78 |
| DESWLsy | 0.5 | 0.85 | 0 | 0.43 | 0.76 |
| WRDMEAc | 0.49 | 0.68 | 0 | 0.7 | 0.63 |
| PCSYNp | 0.48 | 1.45 | 0 | 0.02 | 0.86 |
| CNCADC | 0.46 | 1.61 | 0 | 0.41 | 0.63 |
| WRDFRQc | 0.42 | 0.3 | 0 | 0.83 | 0.45 |
| WRDCNCc | 0.39 | 1.3 | 0 | 0.39 | 0.53 |
| DESWLsyd | 0.39 | 0.32 | 0 | 0.33 | 0.63 |
| SMINTEr | 0.26 | 0.37 | 0 | 0.11 | 0.44 |
| PCCONNz | 0.23 | 0.53 | 0 | 0.23 | 0.32 |
| PCREFp | 0.23 | 0.32 | 0 | 0.01 | 0.44 |
| WRDFAMc | 0.1 | 0 | 0 | 0.24 | 0.09 |
| CRFNO1 | 0.09 | 0.98 | 0 | 0.1 | 0.08 |
| PCCONNp | 0.08 | 1.18 | 0 | 0.04 | 0.06 |
| WRDPRP3p | 0.02 | 0.44 | 0 | 0.03 | 0 |
Coh-Metrix Model 3
General Description
Coh-Metrix Model 3, recommended for current use, is an ensemble (formed by averaging predicted quality scores) of three genre-specific models, detailed below.
The models were trained on Coh-Metrix scores from 15 min narrative, expository, and persuasive writing samples from students in Grades 2-5 to predict holistic writing quality on the samples (theta scores calculated from paired comparisons).
Highly correlated CohMetrix metrics (r > |.90|) were excluded during pre-processing (see section on Scoring Model Development for more details).
More details on the sample will be provided once peer review is complete on the main study using this model.
CohMetrix Model 3narr
This model was trained on CohMetrix scores from 15 min narrative writing samples.
Algorithm Weightings in Ensemble
Abbreviations: * overall = ensemble model * pls = partial least squares regression * mars = bagged multivariate adaptive regression splines * enet = elastic net regression * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | mars | gbm | enet | cube |
|---|---|---|---|---|---|
| 0.0000 | 0.1419 | 0.3143 | 0.0729 | 0.0816 | 0.1792 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | gbm | mars | enet | cube |
|---|---|---|---|---|---|---|
| DESWC | 24.87 | 4.56 | 36.54 | 23.26 | 28.45 | 14.11 |
| WRDHYPn | 8.87 | 1.84 | 2.7 | 14.41 | 7.18 | 4.52 |
| WRDNOUN | 7.12 | 2.27 | 2.89 | 10.61 | 7.1 | 4.37 |
| DESSL | 7 | 1.2 | 0.27 | 14.41 | 0 | 0.77 |
| SYNNP | 5.71 | 1.81 | 3.49 | 7.61 | 8.68 | 2.38 |
| DESWLlt | 5.25 | 0.86 | 1.94 | 7.61 | 5.68 | 4.14 |
| LDVOCD | 4.17 | 2.88 | 3.99 | 6.34 | 0 | 0.69 |
| LDTTRa | 2.6 | 3.58 | 4.54 | 0 | 7.74 | 3.99 |
| SMCAUSwn | 2.18 | 2 | 5.72 | 0 | 3.96 | 2.15 |
| SYNLE | 2.05 | 0.5 | 1.59 | 3.39 | 0 | 0.31 |
| WRDPRP1s | 1.71 | 0.5 | 0.83 | 2.89 | 0 | 0.84 |
| WRDHYPnv | 1.55 | 0.44 | 0.28 | 2.61 | 0 | 1.61 |
| PCDCp | 1.44 | 1.37 | 0.04 | 2.61 | 0.04 | 1 |
| PCREFp | 1.41 | 0.55 | 0 | 2.65 | 0 | 1 |
| PCNARz | 1.32 | 2.01 | 3.19 | 0 | 0 | 3.14 |
| CRFANPa | 1.3 | 1.59 | 3.69 | 0 | 0 | 2.3 |
| LSAGN | 1.29 | 2.26 | 2.08 | 0 | 2.54 | 2.99 |
| WRDFRQmc | 0.94 | 1.95 | 0.78 | 0 | 2.77 | 2.68 |
| CNCLogic | 0.91 | 0.73 | 0.37 | 1.61 | 0 | 0.23 |
| PCDCz | 0.91 | 1.12 | 3.1 | 0 | 0 | 0.77 |
| SYNMEDpos | 0.9 | 1.78 | 0.13 | 0 | 4.2 | 2.61 |
| SMCAUSlsa | 0.84 | 0.78 | 0.76 | 0 | 1.34 | 3.37 |
| PCCONNz | 0.75 | 1.46 | 1.11 | 0 | 2.09 | 1.46 |
| DRPP | 0.65 | 1.2 | 0.36 | 0 | 2.51 | 1.76 |
| WRDAOAc | 0.65 | 1.77 | 1.13 | 0 | 1.24 | 1.23 |
| WRDPRO | 0.63 | 1.35 | 1.42 | 0 | 0 | 1.53 |
| DESPL | 0.61 | 2.71 | 0.51 | 0 | 1.6 | 1.46 |
| PCREFz | 0.59 | 1.06 | 0.56 | 0 | 2.75 | 0.92 |
| WRDMEAc | 0.53 | 0.99 | 1.17 | 0 | 0.82 | 0.84 |
| LDTTRc | 0.52 | 2.28 | 0.15 | 0 | 0 | 2.68 |
| DRNP | 0.49 | 0.62 | 0.24 | 0 | 1.25 | 1.92 |
| DESWLsy | 0.49 | 1.25 | 0.33 | 0 | 0.72 | 1.99 |
| WRDPOLc | 0.44 | 1.45 | 0.91 | 0 | 0 | 1.07 |
| SMINTEr | 0.4 | 0.35 | 0.26 | 0 | 2.03 | 0.84 |
| PCSYNz | 0.39 | 0.91 | 0.59 | 0 | 0 | 1.46 |
| PCCONNp | 0.37 | 2.07 | 0.2 | 0 | 1.9 | 0.31 |
| SMINTEp | 0.35 | 0.56 | 1.24 | 0 | 0 | 0.15 |
| DRPVAL | 0.34 | 1.5 | 0.33 | 0 | 1.65 | 0.23 |
| LSASS1d | 0.33 | 1.49 | 0.06 | 0 | 0 | 1.76 |
| PCCNCz | 0.32 | 1.44 | 0.1 | 0 | 0 | 1.61 |
| CRFCWOa | 0.3 | 1.73 | 0.04 | 0 | 0 | 1.46 |
| RDL2 | 0.29 | 1.91 | 0.31 | 0 | 0 | 0.92 |
| CNCPos | 0.28 | 0.22 | 0.87 | 0 | 0 | 0.38 |
| PCVERBz | 0.28 | 1.5 | 0.14 | 0 | 0 | 1.23 |
| LSAGNd | 0.28 | 1.86 | 0.19 | 0 | 0 | 1.07 |
| CRFCWO1d | 0.25 | 1.57 | 0.74 | 0 | 0 | 0 |
| WRDFRQa | 0.25 | 0.44 | 0.67 | 0 | 0 | 0.46 |
| CNCCaus | 0.25 | 0.36 | 0.29 | 0 | 0 | 1.15 |
| CRFAOa | 0.24 | 1.74 | 0.04 | 0 | 0 | 1.07 |
| DESWLltd | 0.23 | 0.2 | 0.49 | 0 | 0 | 0.69 |
| WRDADJ | 0.22 | 0.25 | 0.43 | 0 | 0 | 0.69 |
| WRDPRP3p | 0.21 | 1.25 | 0.51 | 0 | 0 | 0.23 |
| LDMTLD | 0.21 | 0.26 | 0.42 | 0 | 0 | 0.69 |
| CRFCWOad | 0.21 | 1.68 | 0.35 | 0 | 0 | 0.38 |
| CNCTemp | 0.2 | 0.55 | 0.06 | 0 | 1.45 | 0.15 |
| WRDIMGc | 0.19 | 0.4 | 0.22 | 0 | 0 | 0.84 |
| DESWLsyd | 0.19 | 0.69 | 0.26 | 0 | 0 | 0.69 |
| WRDVERB | 0.18 | 0.55 | 0.44 | 0 | 0 | 0.31 |
| CRFNOa | 0.15 | 1.06 | 0.07 | 0 | 0 | 0.61 |
| CNCADC | 0.14 | 1.46 | 0.12 | 0 | 0 | 0.31 |
| WRDHYPv | 0.14 | 0.35 | 0.32 | 0 | 0 | 0.31 |
| WRDFRQc | 0.14 | 0.58 | 0.38 | 0 | 0 | 0.15 |
| WRDCNCc | 0.14 | 0.37 | 0.51 | 0 | 0 | 0 |
| LSASSp | 0.14 | 1.54 | 0.08 | 0 | 0 | 0.38 |
| LSASSpd | 0.14 | 1.55 | 0.05 | 0 | 0 | 0.46 |
| PCTEMPp | 0.13 | 1.36 | 0.06 | 0 | 0 | 0.38 |
| DRVP | 0.12 | 0.92 | 0.15 | 0 | 0 | 0.31 |
| WRDPRP2 | 0.12 | 1.54 | 0.13 | 0 | 0.23 | 0 |
| DRGERUND | 0.12 | 0.49 | 0.15 | 0 | 0 | 0.46 |
| SMCAUSvp | 0.11 | 0.54 | 0.28 | 0 | 0 | 0.15 |
| PCSYNp | 0.1 | 0.68 | 0.15 | 0 | 0 | 0.23 |
| DRINF | 0.1 | 0.68 | 0.11 | 0 | 0 | 0.31 |
| DRAP | 0.09 | 0.43 | 0.29 | 0 | 0 | 0 |
| WRDADV | 0.09 | 0.6 | 0.28 | 0 | 0 | 0 |
| DESSLd | 0.09 | 1 | 0.12 | 0 | 0.1 | 0.08 |
| SYNSTRUTa | 0.08 | 1.31 | 0.09 | 0 | 0 | 0 |
| PCCNCp | 0.07 | 1.21 | 0 | 0 | 0 | 0.15 |
| WRDFAMc | 0.07 | 0.63 | 0.2 | 0 | 0 | 0 |
| SMCAUSv | 0.06 | 0.53 | 0.14 | 0 | 0 | 0 |
| CNCTempx | 0.05 | 0 | 0.16 | 0 | 0 | 0.08 |
| SMCAUSr | 0.04 | 0.77 | 0.02 | 0 | 0 | 0 |
| PCVERBp | 0.04 | 0.9 | 0 | 0 | 0 | 0 |
| WRDPRP1p | 0.03 | 0.68 | 0.02 | 0 | 0 | 0 |
| WRDPRP3s | 0.02 | 0.4 | 0 | 0 | 0 | 0 |
| DRNEG | 0.01 | 0.24 | 0.02 | 0 | 0 | 0 |
Coh-Metrix Model 3exp
This model was trained on Coh-Metrix scores from 15 min expository writing samples.
Algorithm Weightings in Ensemble
Abbreviations: * overall = ensemble model * pls = partial least squares regression * gbm = stochastic gradient boosted trees * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | mars | gbm | cube |
|---|---|---|---|---|
| -0.0577 | 0.1306 | 0.3136 | 0.3991 | 0.1752 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | mars | pls | gbm | cube |
|---|---|---|---|---|---|
| DESWC | 26.13 | 25.33 | 5.56 | 47.08 | 15.82 |
| LSAGN | 3.77 | 0 | 2.75 | 5.56 | 4.35 |
| LDTTRa | 3.68 | 0 | 4.12 | 4.55 | 3.68 |
| DESSLd | 3.13 | 9.32 | 1.26 | 2.42 | 3.51 |
| DESWLlt | 2.98 | 10.88 | 0.99 | 1.35 | 4.35 |
| WRDPRP2 | 2.25 | 0 | 2.09 | 2.72 | 3.18 |
| LDVOCD | 2.16 | 2.46 | 3.82 | 0.71 | 2.26 |
| DRPP | 2.11 | 0 | 2.07 | 3.28 | 1.09 |
| WRDPOLc | 2.1 | 12.49 | 0.93 | 0.31 | 0.5 |
| LDTTRc | 1.96 | 0 | 3.31 | 1.89 | 1.17 |
| SMCAUSwn | 1.62 | 0 | 0.98 | 2.12 | 2.85 |
| WRDNOUN | 1.61 | 3.23 | 1.38 | 0.53 | 3.26 |
| WRDPRP1s | 1.55 | 5.37 | 1.05 | 0.83 | 1.26 |
| WRDPRP1p | 1.5 | 5.2 | 0.08 | 0.9 | 2.68 |
| CNCTemp | 1.39 | 7.18 | 1.01 | 0.27 | 0.33 |
| PCNARz | 1.37 | 0 | 2.21 | 0.63 | 2.59 |
| LSASS1d | 1.29 | 4.14 | 1.67 | 0.51 | 0.25 |
| PCREFz | 1.27 | 5.37 | 1.14 | 0.18 | 0.92 |
| PCCONNz | 1.25 | 0 | 1.16 | 2.11 | 0.42 |
| DRNP | 1.22 | 3.67 | 0.17 | 1.18 | 1.34 |
| WRDMEAc | 1.2 | 5.37 | 0.64 | 0.47 | 0.75 |
| WRDFRQa | 1.19 | 0 | 1.04 | 1.52 | 1.59 |
| PCCONNp | 1.19 | 0 | 2.2 | 0.62 | 1.59 |
| SYNMEDpos | 1.19 | 0 | 1.84 | 0.24 | 3.1 |
| WRDHYPn | 1.18 | 0 | 0.89 | 1.18 | 2.59 |
| DESPL | 1.03 | 0 | 2.71 | 0.38 | 0.25 |
| WRDHYPnv | 1.02 | 0 | 0.83 | 0.73 | 2.76 |
| PCCNCz | 0.94 | 0 | 1.35 | 0.26 | 2.43 |
| RDL2 | 0.93 | 0 | 1.89 | 0.83 | 0.17 |
| LSASSp | 0.9 | 0 | 1.68 | 0.69 | 0.67 |
| PCVERBz | 0.9 | 0 | 1.38 | 0.38 | 1.92 |
| WRDHYPv | 0.87 | 0 | 0.92 | 0.94 | 1.26 |
| LSASSpd | 0.84 | 0 | 1.66 | 0.21 | 1.42 |
| WRDADJ | 0.82 | 0 | 1.39 | 0.89 | 0.25 |
| CRFCWOa | 0.81 | 0 | 1.79 | 0.37 | 0.67 |
| CRFANPa | 0.78 | 0 | 1.28 | 0.52 | 1.09 |
| LSAGNd | 0.77 | 0 | 2.06 | 0.08 | 0.59 |
| PCREFp | 0.77 | 0 | 0.97 | 0 | 2.76 |
| CRFAOa | 0.74 | 0 | 1.87 | 0.02 | 0.92 |
| DESSL | 0.74 | 0 | 0.94 | 0.46 | 1.59 |
| CRFCWO1d | 0.69 | 0 | 1.77 | 0.29 | 0.17 |
| SYNNP | 0.69 | 0 | 1.25 | 0.31 | 1.09 |
| CRFNOa | 0.64 | 0 | 1.2 | 0.47 | 0.5 |
| PCTEMPp | 0.62 | 0 | 1.26 | 0.49 | 0.25 |
| WRDAOAc | 0.61 | 0 | 1.21 | 0.47 | 0.33 |
| CNCNeg | 0.6 | 0 | 1.7 | 0.2 | 0 |
| DRAP | 0.58 | 0 | 1.08 | 0.21 | 0.92 |
| DRGERUND | 0.57 | 0 | 0.97 | 0.69 | 0 |
| PCDCz | 0.55 | 0 | 1.33 | 0.16 | 0.42 |
| PCDCp | 0.53 | 0 | 1.35 | 0.09 | 0.42 |
| DESWLsy | 0.53 | 0 | 0.58 | 0.34 | 1.26 |
| PCSYNz | 0.51 | 0 | 0.75 | 0.12 | 1.34 |
| DESWLltd | 0.5 | 0 | 0.25 | 0.53 | 1.26 |
| SMCAUSr | 0.47 | 0 | 1.38 | 0.11 | 0 |
| WRDFRQmc | 0.47 | 0 | 1.21 | 0.11 | 0.33 |
| WRDIMGc | 0.45 | 0 | 0.32 | 0.27 | 1.42 |
| LDMTLD | 0.45 | 0 | 0.51 | 0.64 | 0.25 |
| SMCAUSlsa | 0.44 | 0 | 0.33 | 0.24 | 1.42 |
| SYNSTRUTa | 0.43 | 0 | 1.13 | 0.22 | 0 |
| WRDADV | 0.42 | 0 | 0.56 | 0.12 | 1.17 |
| CNCTempx | 0.37 | 0 | 1.07 | 0.09 | 0 |
| WRDFRQc | 0.37 | 0 | 0.68 | 0.16 | 0.59 |
| CNCLogic | 0.36 | 0 | 0.95 | 0.17 | 0 |
| SMINTEr | 0.36 | 0 | 1.12 | 0.03 | 0 |
| DRINF | 0.36 | 0 | 0.89 | 0.21 | 0 |
| DESWLsyd | 0.36 | 0 | 0.28 | 0.55 | 0.33 |
| SMINTEp | 0.35 | 0 | 0.99 | 0.07 | 0.08 |
| SMCAUSvp | 0.33 | 0 | 0.99 | 0.06 | 0 |
| CNCPos | 0.33 | 0 | 0.17 | 0.61 | 0.25 |
| PCVERBp | 0.31 | 0 | 0.49 | 0.01 | 0.92 |
| WRDPRP3s | 0.29 | 0 | 0.42 | 0.26 | 0.33 |
| PCCNCp | 0.29 | 0 | 0.91 | 0.03 | 0 |
| WRDPRO | 0.29 | 0 | 0.64 | 0.14 | 0.25 |
| WRDFAMc | 0.26 | 0 | 0.55 | 0.24 | 0 |
| DRVP | 0.25 | 0 | 0.57 | 0.18 | 0 |
| SMCAUSv | 0.23 | 0 | 0.69 | 0.04 | 0 |
| SYNLE | 0.2 | 0 | 0.03 | 0.33 | 0.33 |
| PCSYNp | 0.2 | 0 | 0.53 | 0.02 | 0.17 |
| WRDPRP3p | 0.19 | 0 | 0.25 | 0.29 | 0 |
| CNCCaus | 0.18 | 0 | 0.47 | 0.09 | 0 |
| WRDVERB | 0.17 | 0 | 0.1 | 0.34 | 0 |
| DRNEG | 0.03 | 0 | 0 | 0.08 | 0 |
Coh-Metrix Model 3per
This model was trained on Coh-Metrix scores from 15 min persuasive writing samples.
Algorithm Weightings in Ensemble
Abbreviations: * overall = ensemble model * pls = partial least squares regression * gbm = stochastic gradient boosted trees * mars = bagged multivariate adaptive regression splines * cube = cubist regression
The table below presents the linear weightings of each algorithm for the ensemble model.
| Intercept | pls | mars | gbm | cube |
|---|---|---|---|---|
| -0.0381 | 0.0558 | 0.4924 | 0.4425 | 0.0259 |
Metric Importance in Each Algorithm and Ensemble
Each column sums to 100 (so values can be interpreted as % contribution to the model).
| Metric | overall | pls | mars | gbm | cube |
|---|---|---|---|---|---|
| DESWC | 32.09 | 4.68 | 34.34 | 33.8 | 19.05 |
| WRDHYPn | 10.41 | 2.03 | 17.13 | 4.3 | 5.17 |
| LDVOCD | 9.59 | 3.43 | 0 | 21.44 | 2.53 |
| DESWLlt | 8.27 | 1.29 | 13.16 | 3.77 | 7.4 |
| LSAGN | 6.13 | 2.92 | 8.63 | 3.88 | 4.05 |
| WRDNOUN | 4.46 | 1.39 | 7.36 | 1.66 | 3.95 |
| WRDADV | 3.05 | 1.18 | 5.53 | 0.52 | 3.24 |
| WRDFRQa | 2.13 | 0.25 | 3.86 | 0.55 | 0.2 |
| SMCAUSwn | 2.11 | 1.08 | 3.7 | 0.54 | 1.01 |
| CNCAdd | 1.76 | 0.66 | 3.37 | 0.19 | 0.2 |
| WRDADJ | 1.67 | 0.59 | 2.93 | 0.26 | 4.15 |
| LDTTRa | 1.47 | 4.05 | 0 | 2.58 | 4.76 |
| DESSC | 1.38 | 3.27 | 0 | 2.6 | 2.74 |
| LDTTRc | 1.17 | 3.17 | 0 | 2.21 | 1.32 |
| DESWLltd | 0.63 | 1.31 | 0 | 1.2 | 1.42 |
| WRDPRO | 0.57 | 1.57 | 0 | 1.1 | 0.3 |
| PCDCz | 0.5 | 1.55 | 0 | 0.94 | 0.3 |
| DESSLd | 0.47 | 1.61 | 0 | 0.84 | 0.61 |
| CRFCWO1d | 0.47 | 2.36 | 0 | 0.74 | 0.81 |
| DRNEG | 0.46 | 1.75 | 0 | 0.72 | 1.82 |
| WRDPOLc | 0.46 | 0.78 | 0 | 0.89 | 1.22 |
| SYNNP | 0.45 | 0.82 | 0 | 0.92 | 0 |
| CNCCaus | 0.44 | 0.24 | 0 | 0.98 | 0 |
| WRDPRP3p | 0.4 | 0.59 | 0 | 0.82 | 0.3 |
| WRDHYPv | 0.39 | 0.94 | 0 | 0.76 | 0.2 |
| LDMTLD | 0.35 | 0.38 | 0 | 0.67 | 1.72 |
| WRDAOAc | 0.34 | 1.9 | 0 | 0.52 | 0.3 |
| CNCPos | 0.32 | 0.48 | 0 | 0.66 | 0 |
| WRDMEAc | 0.32 | 1.11 | 0 | 0.56 | 0.61 |
| CRFANPa | 0.31 | 1.37 | 0 | 0.52 | 0.2 |
| DRVP | 0.31 | 0.47 | 0 | 0.58 | 1.22 |
| WRDFRQc | 0.31 | 0.22 | 0 | 0.65 | 0.71 |
| SMCAUSlsa | 0.26 | 0.65 | 0 | 0.5 | 0 |
| LSASS1d | 0.26 | 2.04 | 0 | 0.34 | 0 |
| CRFAO1 | 0.25 | 2.03 | 0 | 0.3 | 0.2 |
| WRDHYPnv | 0.25 | 0.33 | 0 | 0.48 | 0.71 |
| CNCTempx | 0.23 | 0.5 | 0 | 0.32 | 2.23 |
| SYNSTRUTa | 0.22 | 0.75 | 0 | 0.39 | 0.41 |
| SYNMEDpos | 0.21 | 2.04 | 0 | 0.14 | 1.42 |
| WRDPRP1s | 0.2 | 0.81 | 0 | 0.26 | 1.62 |
| PCVERBz | 0.2 | 1.75 | 0 | 0.15 | 1.62 |
| CRFCWO1 | 0.19 | 1.81 | 0 | 0.18 | 0.41 |
| RDL2 | 0.19 | 1.54 | 0 | 0.22 | 0.51 |
| PCNARz | 0.18 | 2.01 | 0 | 0.05 | 1.72 |
| LSAGNd | 0.18 | 2.08 | 0 | 0.1 | 0.91 |
| RDFKGL | 0.17 | 0.52 | 0 | 0.19 | 2.13 |
| PCSYNz | 0.17 | 0.67 | 0 | 0.19 | 2.13 |
| LSASSpd | 0.15 | 1.95 | 0 | 0.08 | 0.2 |
| SMCAUSv | 0.15 | 0.37 | 0 | 0.26 | 0.61 |
| SYNLE | 0.14 | 0.44 | 0 | 0.26 | 0 |
| RDFRE | 0.14 | 0.45 | 0 | 0.14 | 2.13 |
| DRNP | 0.14 | 0.41 | 0 | 0.28 | 0 |
| PCDCp | 0.14 | 1.89 | 0 | 0 | 1.62 |
| SMCAUSr | 0.13 | 1.29 | 0 | 0.13 | 0 |
| LSASS1 | 0.13 | 1.84 | 0 | 0.04 | 0.41 |
| CRFCWOad | 0.13 | 1.93 | 0 | 0.04 | 0.3 |
| WRDPRP2 | 0.12 | 1.32 | 0 | 0.1 | 0 |
| WRDFAMc | 0.12 | 0.32 | 0 | 0.23 | 0.1 |
| PCREFz | 0.12 | 1.13 | 0 | 0.06 | 1.42 |
| DESWLsy | 0.11 | 0.61 | 0 | 0.13 | 0.51 |
| CNCLogic | 0.11 | 0.55 | 0 | 0.17 | 0.1 |
| CRFNO1 | 0.11 | 1.61 | 0 | 0.04 | 0.2 |
| DRGERUND | 0.1 | 0.38 | 0 | 0.16 | 0.41 |
| DRPP | 0.1 | 0.34 | 0 | 0.19 | 0 |
| PCTEMPp | 0.1 | 1.16 | 0 | 0.07 | 0.2 |
| SMINTEr | 0.1 | 1.5 | 0 | 0.03 | 0.2 |
| PCCNCz | 0.1 | 1.31 | 0 | 0.05 | 0.41 |
| DESWLsyd | 0.1 | 0.76 | 0 | 0.13 | 0.3 |
| CNCNeg | 0.09 | 0.55 | 0 | 0.13 | 0.3 |
| WRDVERB | 0.08 | 0.23 | 0 | 0.15 | 0 |
| SMCAUSvp | 0.08 | 0.24 | 0 | 0.14 | 0.2 |
| PCCONNz | 0.08 | 0.49 | 0 | 0.11 | 0.3 |
| PCCONNp | 0.07 | 1.11 | 0 | 0.01 | 0 |
| DRAP | 0.07 | 0.12 | 0 | 0.14 | 0 |
| WRDCNCc | 0.07 | 0.03 | 0 | 0.14 | 0.2 |
| WRDFRQmc | 0.07 | 1.06 | 0 | 0.03 | 0 |
| PCVERBp | 0.07 | 1.19 | 0 | 0 | 0.3 |
| PCREFp | 0.06 | 0.92 | 0 | 0 | 0.3 |
| PCCNCp | 0.05 | 0.83 | 0 | 0 | 0 |
| WRDPRP1p | 0.05 | 0.16 | 0 | 0.09 | 0 |
| WRDIMGc | 0.05 | 0 | 0 | 0.09 | 0.51 |
| SMINTEp | 0.05 | 0.71 | 0 | 0.03 | 0 |
| DRINF | 0.05 | 0.3 | 0 | 0.06 | 0.3 |
| CNCADC | 0.05 | 0.44 | 0 | 0.05 | 0.2 |
| CNCTemp | 0.04 | 0.58 | 0 | 0.02 | 0 |
| PCSYNp | 0.04 | 0.42 | 0 | 0.01 | 0.71 |
| DRPVAL | 0.01 | 0.07 | 0 | 0.01 | 0 |
Automated Written Expression CBM (aWE-CBM) Model 1
General Description
Total Words Written(TWW) scores are generated directly from the GAMET word count score. Words Spelled Correctly (WSC) scores are generated by subtracting the GAMET misspelling score from the GAMET word count score.
Correct Word Sequences (CWS) and Correct Minus Incorrect Word Sequences (CIWS) scores are based on emsemble models originally trained to predict CBM scores on 7 min narrative writing samples (“I once had a magic pencil and …”) from students in the fall, winter, and spring of Grades 2-5 (Mercer et al., 2019). More details on the sample are available in (Mercer et al., 2019).
The CWS and CIWS models are detailed below (from Mercer et al., 2021).
Correct Word Sequences Model
| Metric | Overall | GBM | SVM | ENET | MARS |
|---|---|---|---|---|---|
| Word Count | 75.48 | 86.79 | 67.10 | 77.17 | 77.84 |
| Spelling | 14.26 | 0.62 | 0.00 | 21.41 | 22.05 |
| %Spelling | 8.78 | 12.28 | 27.95 | 0.40 | 0.11 |
| Grammar | 0.85 | 0.05 | 2.77 | 0.11 | 0.00 |
| %Grammar | 0.01 | 0.06 | 0.01 | 0.00 | 0.00 |
| Duplication | 0.04 | 0.12 | 0.12 | 0.00 | 0.00 |
| Typography | 0.38 | 0.08 | 1.33 | 0.00 | 0.00 |
| White Space | 0.20 | 0.00 | 0.71 | 0.92 | 0.00 |
Note. The weightings sum to 100; thus, they can be viewed as the percentage contribution of each metric to the predicted scores. Overall = the ensemble model of all algorithms, GBM = stochastic gradient boosted regression trees, SVM = support vector machines (radial kernel), ENET = elastic net regression, MARS = bagged multivariate adaptive regression splines. The following regression equation was used to weight the algorithms in the CWS ensemble model: .162 + .074 * GBM + .281 * SVM + .001 * ENET + .642 * MARS.
Correct Minus Incorrect Word Sequences Model
| Metric | Overall | GBM | SVM | ENET | MARS |
|---|---|---|---|---|---|
| Word Count | 55.60 | 55.76 | 47.57 | 61.43 | 61.35 |
| Spelling | 19.25 | 1.48 | 6.57 | 35.80 | 35.04 |
| %Spelling | 22.31 | 41.99 | 42.74 | 0.00 | 0.00 |
| Grammar | 0.82 | 0.00 | 1.69 | 0.00 | 0.62 |
| %Grammar | 0.04 | 0.23 | 0.00 | 0.00 | 0.00 |
| Duplication | 0.28 | 0.10 | 0.76 | 0.00 | 0.00 |
| Typography | 1.37 | 0.41 | 0.07 | 1.55 | 2.97 |
| White Space | 0.34 | 0.04 | 0.60 | 1.22 | 0.00 |
Note. The weightings sum to 100; thus, they can be viewed as the percentage contribution of each metric to the predicted scores. Overall = the ensemble model of all algorithms, GBM = stochastic gradient boosted regression trees, SVM = support vector machines (radial kernel), ENET = elastic net regression, MARS = bagged multivariate adaptive regression splines. The following equation was used for the CIWS model: -.170 + .180 * GBM + .346 * SVM + .100 * ENET + .375 * MARS.